Ribosomal DNA
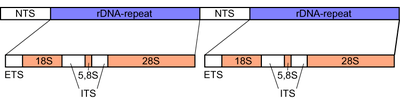
Ribosomal DNA (rDNA) is a DNA sequence that codes for ribosomal RNA. Ribosomes are assemblies of proteins and rRNA molecules that translate mRNA molecules to produce proteins. As shown in the figure, rDNA of eukaryotes consists of a tandem repeat of a unit segment, an operon, composed of NTS, ETS, 18S, ITS1, 5.8S, ITS2, and 28S tracts. rDNA has another gene, coding for 5S rRNA, located in the genome in most eukaryotes.[1] 5S rDNA is also present in tandem repeats as in Drosophila.[1] In the nucleus, the rDNA region of the chromosome is visualized as a nucleolus which forms expanded chromosomal loops with rDNA. These rDNA regions are also called nucleolus organizer regions, as they give rise to the nucleolus. In the human genome there are 5 chromosomes with nucleolus organizer regions: the acrocentric chromosomes 13 (RNR1), 14 (RNR2), 15 (RNR3), 21 (RNR4) and 22 (RNR5). In Bacteria, Archaea and chloroplasts the rRNA is composed of different (smaller) units, the large (23S) ribosomal RNA, 16S ribosomal RNA and 5S rRNA. The 16S rRNA is widely used for phylogenetic studies.[2]
Sequence homogeneity of the repeat unit
In the large rDNA array, polymorphisms between rDNA repeat units are very low, indicating that rDNA tandem arrays are evolving through concerted evolution.[1] However, the mechanism of concerted evolution is imperfect, such that polymorphisms between repeats within an individual can occur at significant levels and may confound phylogenetic analyses for closely related organisms.[3][4]
5S tandem repeat sequences in several Drosophila were compared with each other; the result revealed that insertions and deletions occurred frequently between species and often flanked by conserved sequences.[5] They could occur by slippage of the newly synthesized strand during DNA replication or by gene conversion.[5]
Sequence divergence to clarify phylogeny
The rDNA transcription tracts have low rate of polymorphism among species, which allows interspecific comparison to elucidate phylogenetic relationship using only a few specimens. Coding regions of rDNA are highly conserved among species but ITS regions are variable due to insertions, deletions, and point mutations. Between remote species as human and frog comparison of sequences at ITS tracts is not appropriate.[6] Conserved sequences at coding regions of rDNA allow comparisons of remote species, even between yeast and human. Human 5.8S rRNA has 75% identity with yeast 5.8S rRNA.[7] In cases for sibling species, comparison of the rDNA segment including ITS tracts among species and phylogenetic analysis are made satisfactorily.[8][9] The different coding regions of the rDNA repeats usually show distinct evolutionary rates. As a result, this DNA can provide phylogenetic information of species belonging to wide systematic levels.[10]
Recombination-stimulating activity
A fragment of yeast rDNA containing the 5S gene, nontranscribed spacer DNA, and part of the 35S gene has localized cis-acting mitotic recombination stimulating activity.[11] This DNA fragment contains a mitotic recombination hotspot referred to as HOT1. HOT1 expresses recombination-stimulating activity when it is inserted into novel locations in the yeast genome. HOT1 includes an RNA polymerase I (PolI) transcription promoter that catalyzes 35S ribosomal rRNA gene transcription. In a PolI defective mutant, the HOT1 hotspot recombination-stimulating activity is abolished. The level of PolI transcription in HOT1 appears to determine the level of recombination[12]
References
- 1 2 3 Richard, G. -F.; Kerrest, A.; Dujon, B. (2008). "Comparative Genomics and Molecular Dynamics of DNA Repeats in Eukaryotes". Microbiology and Molecular Biology Reviews. 72 (4): 686–727. doi:10.1128/MMBR.00011-08. PMC 2593564. PMID 19052325.
- ↑ Weisburg WG, Barns SM, Pelletier DA, Lane DJ (January 1991). "16S ribosomal DNA amplification for phylogenetic study". J. Bacteriol. 173 (2): 697–703. doi:10.1128/jb.173.2.697-703.1991. PMC 207061. PMID 1987160.
- ↑ Álvarez, I.; Wendel, J. F. (2003). "Ribosomal ITS sequences and plant phylogenetic inference". Molecular Phylogenetics and Evolution. 29 (3): 417–434. doi:10.1016/S1055-7903(03)00208-2. PMID 14615184.
- ↑ Weitemier, K; Straub, S. C. K.; Fishbein, M.; Liston, A. (2015). "Intragenomic polymorphisms among high-copy loci: a genus-wide study of nuclear ribosomal DNA in Asclepias (Apocynaceae)". PeerJ. 3: e718. doi:10.7717/peerj.718. PMC 4304868. PMID 25653903.
- 1 2 Päques, F.; Samson, M. L.; Jordan, P.; Wegnez, M. (1995). "Structural evolution of the Drosophila 5S ribosomal genes". Journal of Molecular Evolution. 41 (5): 615–621. doi:10.1007/bf00175820. PMID 7490776.
- ↑ Sumida, M.; Kato, Y.; Kurabayashi, A. (2004). "Sequencing and analysis of the internal transcribed spacers (ITSs) and coding regions in the EcoR I fragment of the ribosomal DNA of the Japanese pond frog Rana nigromaculata". Genes & genetic systems. 79 (2): 105–118. doi:10.1266/ggs.79.105. PMID 15215676.
- ↑ Nazar, R. N.; Sitz, T. O.; Busch, H. (1976). "Sequence homologies in mammalian 5.8S ribosomal RNA". Biochemistry. 15 (3): 505–508. doi:10.1021/bi00648a008. PMID 1252408.
- ↑ Ma, YJ; Qu, FY; Xu, JJ (1998). "Sequence differences of rDNA-ITS2 and species-diagnostic PCR assay of Anopheles sinensis and Anopheles anthropophagus from China" (PDF). J Med Coll PLA. 13: 123–128.
- ↑ Li, C; Lee, JS; Groebner, JL; Kim, HC; Klein, TA; O'Guinn, ML; Wilkerson, RC (2005). "A newly recognized species in the Anopheles hyrcanus group and molecular identification of related species from the Republic of South Korea (Diptera: Culicidae)". Zootaxa. 939: 1–8. doi:10.11646/zootaxa.939.1.1.
- ↑ Hillis, D. M.; Dixon, M. T. (1991). "Ribosomal DNA: Molecular evolution and phylogenetic inference". The Quarterly Review of Biology. 66 (4): 411–453. doi:10.1086/417338. PMID 1784710.
- ↑ Keil RL, Roeder GS (1984). "Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae". Cell. 39 (2 Pt 1): 377–86. doi:10.1016/0092-8674(84)90016-3. PMID 6094015.
- ↑ Serizawa N, Horiuchi T, Kobayashi T (2004). "Transcription-mediated hyper-recombination in HOT1". Genes Cells. 9 (4): 305–15. doi:10.1111/j.1356-9597.2004.00729.x. PMID 15066122.