TxpA-RatA toxin-antitoxin system
| RNA anti-toxin A | |
|---|---|
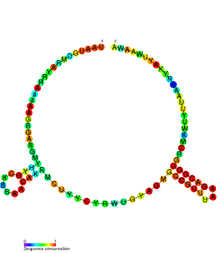 Conserved secondary structure of RatA antitoxin sRNA. | |
| Identifiers | |
| Symbol | RatA |
| Rfam | RF01776 |
| Other data | |
| RNA type | Antisense |
| Domain(s) | Firmicutes |
| SO | {{{SO}}} |
| PDB structures | PDBe |
The TxpA/RatA toxin-antitoxin system was first identified in Bacillus subtilis.[1] It consists of a non-coding 222nt sRNA called RatA (RNA anti-toxin A) and a protein toxin named TxpA (Toxic protein A).[2]
RatA was discovered in intergenic regions of the B. subtilis genome, in a 728-nucleotide region between genes yqdB (later renamed TxpA) and yqbM. Initially, Affymetrix microarrays were used to detect transcription in this region,[3] Northern blot experiments and mutation analysis were then employed to characterise the RNA transcript.[1]
ratA and txpA are transcribed convergently, and have an overlap of 75 base pairs at their 3' ends, providing an area of complimentarity. The RatA transcript binds with the TxpA mRNA across the complimentary region[1] and the dsRNA is then degraded by an uncharacterised RNase E equivalent, preventing translation of the toxic TxpA protein.[2]
Genes ratA and txpA are found within a 48kb phage-like element called skin. This element interrupts a gene for the sigma factor σK and is excised during sporulation. The toxin-antitoxin system contained within skin forces the inheritance of this element, which is acting as a selfish gene.[1]
The mechanism by which TxpA induces cell lysis and death is unknown. TxpA is not similar enough to other proteins of known function to infer a related function, however it does have a suspected transmembrane region in its N-terminal, so it is possible that TxpA damages the integrity of the cell membrane, or blocks cell wall synthesis.[1]
See also
References
- 1 2 3 4 5 Silvaggi JM, Perkins JB, Losick R (October 2005). "Small untranslated RNA antitoxin in Bacillus subtilis". J. Bacteriol. 187 (19): 6641–50. doi:10.1128/JB.187.19.6641-6650.2005. PMC 1251590. PMID 16166525. Retrieved 2010-09-16.
- 1 2 Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G (June 2010). "Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families". Nucleic Acids Res. 38 (11): 3743–59. doi:10.1093/nar/gkq054. PMC 2887945. PMID 20156992. Retrieved 2010-09-16.
- ↑ Lee JM, Zhang S, Saha S, Santa Anna S, Jiang C, Perkins J (December 2001). "RNA expression analysis using an antisense Bacillus subtilis genome array". J. Bacteriol. 183 (24): 7371–80. doi:10.1128/JB.183.24.7371-7380.2001. PMC 95586. PMID 11717296. Retrieved 2010-09-16.
Further reading
- Gerdes K, Wagner EG (April 2007). "RNA antitoxins". Curr. Opin. Microbiol. 10 (2): 117–24. doi:10.1016/j.mib.2007.03.003. PMID 17376733. Retrieved 2010-08-10.
- Pandey DP, Gerdes K (2005). "Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes". Nucleic Acids Res. 33 (3): 966–76. doi:10.1093/nar/gki201. PMC 549392. PMID 15718296. Retrieved 2010-08-11.
- Hayes F (September 2003). "Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest". Science. 301 (5639): 1496–9. doi:10.1126/science.1088157. PMID 12970556. Retrieved 2010-08-11.
- Fozo EM, Hemm MR, Storz G (December 2008). "Small toxic proteins and the antisense RNAs that repress them". Microbiol. Mol. Biol. Rev. 72 (4): 579–89, Table of Contents. doi:10.1128/MMBR.00025-08. PMC 2593563. PMID 19052321. Retrieved 2010-08-11.
- Mochizuki A, Yahara K, Kobayashi I, Iwasa Y (February 2006). "Genetic addiction: selfish gene's strategy for symbiosis in the genome". Genetics. 172 (2): 1309–23. doi:10.1534/genetics.105.042895. PMC 1456228. PMID 16299387. Retrieved 2010-08-13.