RNA polymerase
| DNA-Directed RNA Polymerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 2.7.7.6 | ||||||||
| CAS number | 9014-24-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
RNA polymerase (ribonucleic acid polymerase), both abbreviated RNAP or RNApol, official name DNA-directed RNA polymerase, is a member of a family of enzymes that are essential to life: they are found in all living organisms and many viruses. RNAP locally opens the double-stranded DNA (usually about four turns of the double helix) so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP has intrinsic helicase activity, therefore no separate enzyme is needed to unwind the DNA (in contrast to DNA polymerase). RNAP not only initiates RNA transcription, it also guides the nucleotides into position, facilitates attachment and elongation, has intrinsic proofreading and replacement capabilities, and termination recognition capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides.
RNAP produces RNA that functionally is either coding (for protein) (messenger RNA) (mRNA); or non-coding: so-called "RNA genes". At least four functional types of RNA genes exist:
- transfer RNA (tRNA) — transfers specific amino acids to growing polypeptide chains at the ribosomal site of protein synthesis during translation;
- ribosomal RNA (rRNA) — incorporates into ribosomes;
- micro RNA (miRNA) — regulates gene activity; and,
- catalytic RNA (ribozyme) — functions as an enzymatically active RNA molecule.
Eukaryotes have multiple types of nuclear RNAP, each responsible for synthesis of a distinct subset of RNA. All are structurally and mechanistically related to each other and to bacterial RNAP. RNA polymerase I synthesizes a pre-rRNA 45S (35S in yeast), which matures and will form the major RNA sections of the ribosome. RNA polymerase II synthesizes precursors of mRNAs and most snRNA and microRNAs. RNA polymerase III synthesizes tRNAs, rRNA 5S and other small RNAs found in the nucleus and cytosol. RNA polymerase IV synthesizes siRNA in plants. RNA polymerase V synthesizes RNAs involved in siRNA-directed heterochromatin formation in plants. Eukaryotic chloroplasts contain an RNAP very highly structurally and mechanistically similar to bacterial RNAP ("plastid-encoded polymerase"). They also contain a second, structurally and mechanistically unrelated, RNAP ("nucleus-encoded polymerase"; member of the "single-subunit RNAP" protein family). Eukaryotic mitochondria contain a structurally and mechanistically unrelated RNAP (member of the "single-subunit RNAP" protein family).
Given that DNA and RNA polymerases both carry out template-dependent nucleotide polymerization, it might be expected that the two types of enzymes would be structurally related, but they are not. They seem to have arisen independently twice during the early evolution of cells: one lineage led to the modern DNA polymerases and reverse transcriptases, as well as to a few single-subunit RNA polymerases from viruses; the other lineage formed all of the modern cellular RNA polymerases.
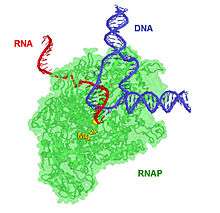
Structure
The 2006 Nobel Prize in Chemistry was awarded to Roger D. Kornberg for creating detailed molecular images of RNA polymerase during various stages of the transcription process.[1] In most prokaryotes, a single RNA polymerase species transcribes all types of RNA. RNA polymerase from E. coli consists of five different subunit types. The beta (β) subunit has a molecular weight of 150,000, beta prime (β′) 160,000, alpha (α) 40,000, and sigma (σ) 70,000. The σ subunit can dissociate from the rest of the complex, leaving the core enzyme. The complete enzyme with σ is termed the RNA polymerase holoenzyme and is necessary for correct initiation of transcription, whereas the core enzyme can continue transcription after initiation.[2]
All RNAPs contain metal cofactors, in particular zinc and magnesium cations which aid in the transcription process.[3][4]
Function
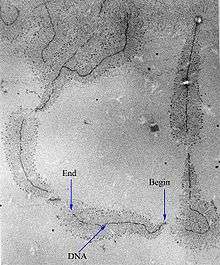
Control of the process of gene transcription affects patterns of gene expression and, thereby, allows a cell to adapt to a changing environment, perform specialized roles within an organism, and maintain basic metabolic processes necessary for survival. Therefore, it is hardly surprising that the activity of RNAP is long, complex, and highly regulated. In Escherichia coli bacteria, more than 100 transcription factors have been identified, which modify the activity of RNAP.[5]
RNAP can initiate transcription at specific DNA sequences known as promoters. It then produces an RNA chain, which is complementary to the template DNA strand. The process of adding nucleotides to the RNA strand is known as elongation; in eukaryotes, RNAP can build chains as long as 2.4 million nucleotides (the full length of the dystrophin gene). RNAP will preferentially release its RNA transcript at specific DNA sequences encoded at the end of genes, which are known as terminators.
Products of RNAP include:
- Messenger RNA (mRNA)—template for the synthesis of proteins by ribosomes.
- Non-coding RNA or "RNA genes"—a broad class of genes that encode RNA that is not translated into protein. The most prominent examples of RNA genes are transfer RNA (tRNA) and ribosomal RNA (rRNA), both of which are involved in the process of translation. However, since the late 1990s, many new RNA genes have been found, and thus RNA genes may play a much more significant role than previously thought.
- Transfer RNA (tRNA)—transfers specific amino acids to growing polypeptide chains at the ribosomal site of protein synthesis during translation
- Ribosomal RNA (rRNA)—a component of ribosomes
- Micro RNA—regulates gene activity
- Catalytic RNA (Ribozyme)—enzymatically active RNA molecules
RNAP accomplishes de novo synthesis. It is able to do this because specific interactions with the initiating nucleotide hold RNAP rigidly in place, facilitating chemical attack on the incoming nucleotide. Such specific interactions explain why RNAP prefers to start transcripts with ATP (followed by GTP, UTP, and then CTP). In contrast to DNA polymerase, RNAP includes helicase activity, therefore no separate enzyme is needed to unwind DNA.
Action
RNA polymerase binding in bacteria involves the sigma factor recognizing the core promoter region containing the -35 and -10 elements (located before the beginning of sequence to be transcribed) and also, at some promoters, the α subunit C-terminal domain recognizing promoter upstream elements. There are multiple interchangeable sigma factors, each of which recognizes a distinct set of promoters. For example, in E. coli, σ70 is expressed under normal conditions and recognizes promoters for genes required under normal conditions ("housekeeping genes"), while σ32 recognizes promoters for genes required at high temperatures ("heat-shock genes").
After binding to the DNA, the RNA polymerase switches from a closed complex to an open complex. This change involves the separation of the DNA strands to form an unwound section of DNA of approximately 13 bp, referred to as the transcription bubble. Ribonucleotides are base-paired to the template DNA strand, according to Watson-Crick base-pairing interactions. Supercoiling plays an important part in polymerase activity because of the unwinding and rewinding of DNA. Because regions of DNA in front of RNAP are unwound, there are compensatory positive supercoils. Regions behind RNAP are rewound and negative supercoils are present.
As noted above, RNA polymerase makes contacts with the promoter region. However these stabilizing contacts inhibit the enzyme's ability to access DNA further downstream and thus the synthesis of the full-length product. Once the open complex is stabilized, RNA polymerase synthesizes an RNA strand to establish a DNA-RNA heteroduplex (~8-9 bp) at the active center, which stabilizes the elongation complex. In order to accomplish RNA synthesis, RNA polymerase must maintain promoter contacts while unwinding more downstream DNA for synthesis, "scrunching" more downstream DNA into the initiation complex. During the promoter escape transition, RNA polymerase is considered a "stressed intermediate." Thermodynamically the stress accumulates from the DNA-unwinding and DNA-compaction activities. Once the DNA-RNA heteroduplex is long enough, RNA polymerase releases its upstream contacts and effectively achieves the promoter escape transition into the elongation phase. However, promoter escape is not the only outcome. RNA polymerase can also relieve the stress by releasing its downstream contacts, arresting transcription. The paused transcribing complex has two options: (1) release the nascent transcript and begin anew at the promoter or (2) reestablish a new 3'OH on the nascent transcript at the active site via RNA polymerase's catalytic activity and recommence DNA scrunching to achieve promoter escape. Scientists have coined the term "abortive initiation" to explain the unproductive cycling of RNA polymerase before the promoter escape transition. The extent of abortive initiation depends on the presence of transcription factors and the strength of the promoter contacts.
Elongation

Transcription elongation involves the further addition of ribonucleotides and the change of the open complex to the transcriptional complex. RNAP cannot start forming full length transcripts because of its strong binding to the promoter. Transcription at this stage primarily results in short RNA fragments of around 9 bp in a process known as abortive transcription. Once the RNAP starts forming longer transcripts it clears the promoter. At this point, the contacts with the -10 and -35 elements are disrupted, and the σ factor falls off RNAP. This allows the rest of the RNAP complex to move forward, as the σ factor held the RNAP complex in place.
The 17-bp transcriptional complex has an 8-bp DNA-RNA hybrid, that is, 8 base-pairs involve the RNA transcript bound to the DNA template strand. As transcription progresses, ribonucleotides are added to the 3' end of the RNA transcript and the RNAP complex moves along the DNA.
Aspartyl (asp) residues in the RNAP will hold on to Mg2+ ions, which will, in turn, coordinate the phosphates of the ribonucleotides. The first Mg2+ will hold on to the α-phosphate of the NTP to be added. This allows the nucleophilic attack of the 3'OH from the RNA transcript, adding another NTP to the chain. The second Mg2+ will hold on to the pyrophosphate of the NTP. The overall reaction equation is:
(NMP)n + NTP --> (NMP)n+1 + PPi
Fidelity
Unlike the proofreading mechanisms of DNA polymerase those of the RNA variety have only recently been investigated. Proofreading begins with separation of the mis-incorporated nucleotide from the DNA template. This pauses transcription. The polymerase then backtracks by one position and cleaves the dinucleotide that contains the mismatched nucleotide. In the RNA polymerase this occurs at the same active site used for polymerization and is therefore markedly different from the DNA polymerase where proofreading occurs at a distinct nuclease active site.[6]
Termination
In prokaryotes, termination of RNA transcription can be rho-independent or rho-dependent:
Rho-independent transcription termination is the termination of transcription without the aid of the rho protein. Transcription of a palindromic region of DNA causes the formation of a "hairpin" structure from the RNA transcription looping and binding upon itself. This hairpin structure is often rich in G-C base-pairs, making it more stable than the DNA-RNA hybrid itself. As a result, the 8 bp DNA-RNA hybrid in the transcription complex shifts to a 4 bp hybrid. These last 4 base pairs are weak A-U base pairs, and the entire RNA transcript will fall off the DNA.
Other organisms
Bacteria
In bacteria, the same enzyme catalyzes the synthesis of mRNA and non-coding RNA (ncRNA).
RNAP is a large molecule. The core enzyme has five subunits (~400 kDa):[7]
- β': The β' subunit is the largest subunit, and is encoded by the rpoC gene.[8] The β' subunit contains part of the active center responsible for RNA synthesis and contains some of the determinants for non-sequence-specific interactions with DNA and nascent RNA.
- β: The β subunit is the second-largest subunit, and is encoded by the rpoB gene. The β subunit contains the rest of the active center responsible for RNA synthesis and contains the rest of the determinants for non-sequence-specific interactions with DNA and nascent RNA.
- αI and αII: The α subunit is the third-largest subunit and is present in two copies per molecule of RNAP, αI and αII. Each α subunit contains two domains: αNTD (N-Terminal domain) and αCTD (C-terminal domain). αNTD contains determinants for assembly of RNAP. αCTD (C-terminal domain) contains determinants for interaction with promoter DNA, making non-sequence-non-specific interactions at most promoters and sequence-specific interactions at upstream-element-containing promoters, and contains determinants for interactions with regulatory factors.
- ω: The ω subunit is the smallest subunit. The ω subunit facilitates assembly of RNAP and stabilizes assembled RNAP.[9]
In order to bind promoters, RNAP core associates with the transcription initiation factor sigma (σ) to form RNA polymerase holoenzyme. Sigma reduces the affinity of RNAP for nonspecific DNA while increasing specificity for promoters, allowing transcription to initiate at correct sites. The complete holoenzyme therefore has 6 subunits: β'βαI and αIIωσ (~450 kDa).
Eukaryotes

Eukaryotes have multiple types of nuclear RNAP, each responsible for synthesis of a distinct subset of RNA. All are structurally and mechanistically related to each other and to bacterial RNAP:
- RNA polymerase I synthesizes a pre-rRNA 45S (35S in yeast), which matures into 28S, 18S and 5.8S rRNAs which will form the major RNA sections of the ribosome.[10]
- RNA polymerase II synthesizes precursors of mRNAs and most snRNA and microRNAs.[11] This is the most studied type, and, due to the high level of control required over transcription, a range of transcription factors are required for its binding to promoters.
- RNA polymerase III synthesizes tRNAs, rRNA 5S and other small RNAs found in the nucleus and cytosol.[12]
- RNA polymerase IV synthesizes siRNA in plants.[13]
- RNA polymerase V synthesizes RNAs involved in siRNA-directed heterochromatin formation in plants.[14]
Eukaryotic chloroplasts contain an RNAP very highly structurally and mechanistically similar to bacterial RNAP ("plastid-encoded polymerase").
Eukaryotic chloroplasts also contain a second, structurally and mechanistically unrelated, RNAP ("nucleus-encoded polymerase"; member of the "single-subunit RNAP" protein family).
Eukaryotic mitochondria contain a structurally and mechanistically unrelated RNAP (member of the "single-subunit RNAP" protein family).
Given that DNA and RNA polymerases both carry out template-dependent nucleotide polymerization, it might be expected that the two types of enzymes would be structurally related. However, x-ray crystallographic studies of both types of enzymes reveal that, other than containing a critical Mg2+ ion at the catalytic site, they are virtually unrelated to each other; indeed template-dependent nucleotide polymerizing enzymes seem to have arisen independently twice during the early evolution of cells. One lineage led to the modern DNA Polymerases and reverse transcriptases, as well as to a few single-subunit RNA polymerases from viruses. The other lineage formed all of the modern cellular RNA polymerases.
Archaea
Archaea have a single type of RNAP, responsible for the synthesis of all RNA. Archaeal RNAP is structurally and mechanistically similar to bacterial RNAP and eukaryotic nuclear RNAP I-V, and is especially closely structurally and mechanistically related to eukaryotic nuclear RNAP II.[15][16] The history of the discovery of the archaeal RNA polymerase is quite recent. The first analysis of the RNAP of an archaeon was performed in 1971, when the RNAP from the extreme halophile Halobacterium cutirubrum was isolated and purified.[17] Crystal structures of RNAPs from Sulfolobus solfataricus and Sulfolobus shibatae set the total number of identified archaeal subunits at thirteen.[15][18]
Viruses

Orthopoxviruses synthesize RNA using a virally encoded RNAP that is structurally and mechanistically related to bacterial RNAP, archaeal RNAP, and eukaryotic nuclear RNAP I-V. Most other viruses that synthesize RNA using a virally encoded RNAP use an RNAP that is not structurally and mechanistically related to bacterial RNAP, archaeal RNAP, and eukaryotic nuclear RNAP I-V. Many viruses use a single-subunit DNA-dependent RNAP that is structurally and mechanistically related to the single-subunit RNAP of eukaryotic chloroplasts and mitochondria and, more distantly, to DNA polymerases and reverse transcriptases. Perhaps the most widely studied such single-subunit RNAP is bacteriophage T7 RNA polymerase. Other viruses use a RNA-dependent RNAP (an RNAP that employs RNA as a template instead of DNA). This occurs in negative strand RNA viruses and dsRNA viruses, both of which exist for a portion of their life cycle as double-stranded RNA. However, some positive strand RNA viruses, such as poliovirus, also contain RNA-dependent RNAP.[19]
History
RNAP was discovered independently by Charles Loe, Audrey Stevens, and Jerard Hurwitz in 1960.[20] By this time, one half of the 1959 Nobel Prize in Medicine had been awarded to Severo Ochoa for the discovery of what was believed to be RNAP,[21] but instead turned out to be polynucleotide phosphorylase.
Purification
RNA polymerase can be isolated in the following ways:
- By a phosphocellulose column.[22]
- By glycerol gradient centrifugation.[23]
- By a DNA column.
- By an ion chromatography column.[24]
And also combinations of the above techniques.
See also
- Alpha-amanitin
- Transcription Activators in Eukaryotes
References
- ↑ Nobel Prize in Chemistry 2006
- ↑ Griffiths AJF, Miller JH, Suzuki DT, et al. An Introduction to Genetic Analysis. 7th edition. New York: W. H. Freeman; 2000. Chapter 10.
- ↑ Bruce,, Alberts,. Molecular biology of the cell (Sixth ed.). New York, NY. ISBN 9780815344322. OCLC 887605755.
- ↑ Markov, Dmitriy; Naryshkina, Tatyana; Mustaev, Arkady; Severinov, Konstantin (1999-09-15). "A zinc-binding site in the largest subunit of DNA-dependent RNA polymerase is involved in enzyme assembly". Genes & Development. 13 (18): 2439–2448. ISSN 0890-9369. PMID 10500100.
- ↑ Akira Ishihama (2000). "Functional modulation of Escherichia coli RNA polymerase". Annu. Rev. Microbiol. 54: 499–518. doi:10.1146/annurev.micro.54.1.499. PMID 11018136.
- ↑ Sydow, Jasmin F.; Cramer, Patrick (December 2009). "RNA polymerase fidelity and transcriptional proofreading" (PDF). Current Opinion in Structural Biology. 19 (6): 732–739. doi:10.1016/j.sbi.2009.10.009. ISSN 1879-033X. PMID 19914059.
- ↑ Ebright RH (2000). "RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II". J Mol Biol. 304 (5): 687–98. doi:10.1006/jmbi.2000.4309. PMID 11124018.
- ↑ Ovchinnikov, Yu; Monastyrskaya, G; Gubanov, V; Guryev, S; Salomatina, I; Shuvaeva, T; Lipkin, V; Sverdlov, E (1982). "The primary structure of E. coli RNA polymerase. Nucleotide sequence of the rpoC gene and amino acid sequence of the β′-subunit". Nucleic Acids Research. 10 (13): 4035–4044. doi:10.1093/nar/10.13.4035. Retrieved 16 November 2014.
- ↑ Mathew, Renjith; Chatterji, Dipankar (October 2006). "The evolving story of the omega subunit of bacterial RNA polymerase". Trends in Microbiology. 14 (10): 450–455. doi:10.1016/j.tim.2006.08.002. Retrieved 17 November 2014.
- ↑ Grummt I. (1999). "Regulation of mammalian ribosomal gene transcription by RNA polymerase I.". Prog Nucleic Acid Res Mol Biol. 62: 109–54. doi:10.1016/S0079-6603(08)60506-1. PMID 9932453.
- ↑ Lee Y; Kim M; Han J; Yeom KH; Lee S; Baek SH; Kim VN. (October 2004). "MicroRNA genes are transcribed by RNA polymerase II". EMBO J. 23 (20): 4051–60. doi:10.1038/sj.emboj.7600385. PMC 524334. PMID 15372072.
- ↑ Willis IM. (February 1993). "RNA polymerase III. Genes, factors and transcriptional specificity". Eur. J. Biochem. 212 (1): 1–11. doi:10.1111/j.1432-1033.1993.tb17626.x. PMID 8444147.
- ↑ Herr AJ, Jensen MB, Dalmay T, Baulcombe DC (2005). "RNA polymerase IV directs silencing of endogenous DNA". Science. 308 (5718): 118–20. doi:10.1126/science.1106910. PMID 15692015.
- ↑ Wierzbicki AT, Ream TS, Haag JR, Pikaard CS (May 2009). "RNA Polymerase V transcription guides ARGONAUTE4 to chromatin". Nat. Genet. 41 (5): 630–4. doi:10.1038/ng.365. PMC 2674513. PMID 19377477.
- 1 2 Korkhin, Y; Unligil, U. M.; Littlefield, O; Nelson, P. J.; Stuart, D. I.; Sigler, P. B.; Bell, S. D.; Abrescia, N. G. (2009). "Evolution of complex RNA polymerases: The complete archaeal RNA polymerase structure". PLoS Biology. 7 (5): e1000102. doi:10.1371/journal.pbio.1000102. PMC 2675907. PMID 19419240.
- ↑ Werner, F (2007). "Structure and function of archaeal RNA polymerases". Molecular Microbiology. 65 (6): 1395–404. doi:10.1111/j.1365-2958.2007.05876.x. PMID 17697097.
- ↑ Louis, B. G.; Fitt, P. S. (1971). "Nucleic acid enzymology of extremely halophilic bacteria. Halobacterium cutirubrum deoxyribonucleic acid-dependent ribonucleic acid polymerase". The Biochemical Journal. 121 (4): 621–7. doi:10.1042/bj1210621. PMC 1176638. PMID 4940048.
- ↑ Hirata, A.; Klein, B.; Murakami, K. (2008). "The X-ray crystal structure of RNA polymerase from Archaea". Nature. 451 (7180): 851–854. doi:10.1038/nature06530. PMC 2805805. PMID 18235446.
- ↑ Ahlquist, Paul (2002). "RNA-Dependent RNA Polymerases, Viruses, and RNA Silencing". Science. 296: 1270–1273. doi:10.1126/science.1069132. PMID 12016304.
- ↑ Jerard Hurwitz (December 2005). "The Discovery of RNA Polymerase". Journal of Biological Chemistry. 280 (52): 42477–85. doi:10.1074/jbc.X500006200. PMID 16230341.
- ↑ Nobel Prize 1959
- ↑ Kelly JL; Lehman IR. (August 1986). "Yeast mitochondrial RNA polymerase. Purification and properties of the catalytic subunit". J Biol Chem. 261 (22): 10340–7. PMID 3525543.
- ↑ Honda A, et al. (April 1990). "Purification and molecular structure of RNA polymerase from influenza virus A/PR8". J Biochem. 107 (4): 624–8. PMID 2358436.
- ↑ Hager; et al. (1990). "Use of Mono Q High-Resolution Ion-Exchange Chromatography To Obtain Highly Pure and Active Escherichia coli RNA Polymerase". Biochemistry. 29 (34): 7890–7894. doi:10.1021/bi00486a016. PMID 2261443.
External links
| Wikimedia Commons has media related to RNA polymerase. |
- DNAi - DNA Interactive, including information and Flash clips on RNA Polymerase.
- RNA+Polymerase at the US National Library of Medicine Medical Subject Headings (MeSH)
- EC 2.7.7.6
- RNA Polymerase - Synthesis RNA from DNA Template
- 3D macromolecular structures of RNA Polymerase from the EM Data Bank(EMDB)