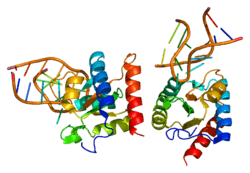NHP2L1
NHP2-like protein 1 is a protein that in humans is encoded by the NHP2L1 gene.[5][6]
Function
Originally named because of its sequence similarity to the Saccharomyces cerevisiae NHP2 (non-histone protein 2), this protein appears to be a highly conserved nuclear protein that is a component of the [U4/U6.U5] tri-snRNP. It binds to the 5' stem-loop of U4 snRNA. Two transcript variants encoding the same protein have been found for this gene.[6]
Interactions
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000100138 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000063480 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Saito H, Fujiwara T, Shin S, Okui K, Nakamura Y (Jan 1997). "Cloning and mapping of a human novel cDNA (NHP2L1) that encodes a protein highly homologous to yeast nuclear protein NHP2". Cytogenetics and Cell Genetics. 72 (2–3): 191–3. doi:10.1159/000134186. PMID 8978773.
- 1 2 "Entrez Gene: NHP2L1 NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae)".
- ↑ Chang MS, Sasaki H, Campbell MS, Kraeft SK, Sutherland R, Yang CY, Liu Y, Auclair D, Hao L, Sonoda H, Ferland LH, Chen LB (Dec 1999). "HRad17 colocalizes with NHP2L1 in the nucleolus and redistributes after UV irradiation". The Journal of Biological Chemistry. 274 (51): 36544–9. doi:10.1074/jbc.274.51.36544. PMID 10593953.
Further reading
- Nottrott S, Hartmuth K, Fabrizio P, Urlaub H, Vidovic I, Ficner R, Lührmann R (Nov 1999). "Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5' stem-loop of U4 snRNA". The EMBO Journal. 18 (21): 6119–33. doi:10.1093/emboj/18.21.6119. PMC 1171676. PMID 10545122.
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, Ainscough R, Almeida JP, Babbage A, Bagguley C, Bailey J, Barlow K, Bates KN, Beasley O, Bird CP, Blakey S, Bridgeman AM, Buck D, Burgess J, Burrill WD, O'Brien KP (Dec 1999). "The DNA sequence of human chromosome 22". Nature. 402 (6761): 489–95. doi:10.1038/990031. PMID 10591208.
- Chang MS, Sasaki H, Campbell MS, Kraeft SK, Sutherland R, Yang CY, Liu Y, Auclair D, Hao L, Sonoda H, Ferland LH, Chen LB (Dec 1999). "HRad17 colocalizes with NHP2L1 in the nucleolus and redistributes after UV irradiation". The Journal of Biological Chemistry. 274 (51): 36544–9. doi:10.1074/jbc.274.51.36544. PMID 10593953.
- Vidovic I, Nottrott S, Hartmuth K, Lührmann R, Ficner R (Dec 2000). "Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment". Molecular Cell. 6 (6): 1331–42. doi:10.1016/S1097-2765(00)00131-3. PMID 11163207.
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI (Jan 2002). "Directed proteomic analysis of the human nucleolus". Current Biology. 12 (1): 1–11. doi:10.1016/S0960-9822(01)00650-9. PMID 11790298.
- Leung AK, Lamond AI (May 2002). "In vivo analysis of NHPX reveals a novel nucleolar localization pathway involving a transient accumulation in splicing speckles". The Journal of Cell Biology. 157 (4): 615–29. doi:10.1083/jcb.200201120. PMC 2173864. PMID 12011111.
- Watkins NJ, Dickmanns A, Lührmann R (Dec 2002). "Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP". Molecular and Cellular Biology. 22 (23): 8342–52. doi:10.1128/MCB.22.23.8342-8352.2002. PMC 134055. PMID 12417735.
- Scherl A, Couté Y, Déon C, Callé A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ (Nov 2002). "Functional proteomic analysis of human nucleolus". Molecular Biology of the Cell. 13 (11): 4100–9. doi:10.1091/mbc.E02-05-0271. PMC 133617. PMID 12429849.
- Collins JE, Wright CL, Edwards CA, Davis MP, Grinham JA, Cole CG, Goward ME, Aguado B, Mallya M, Mokrab Y, Huckle EJ, Beare DM, Dunham I (2005). "A genome annotation-driven approach to cloning the human ORFeome". Genome Biology. 5 (10): R84. doi:10.1186/gb-2004-5-10-r84. PMC 545604. PMID 15461802.
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M (Jan 2005). "Nucleolar proteome dynamics". Nature. 433 (7021): 77–83. doi:10.1038/nature03207. PMID 15635413.
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE (Sep 2005). "A human protein-protein interaction network: a resource for annotating the proteome". Cell. 122 (6): 957–68. doi:10.1016/j.cell.2005.08.029. PMID 16169070.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (Oct 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. doi:10.1038/nature04209. PMID 16189514.
This article is issued from
Wikipedia.
The text is licensed under Creative Commons - Attribution - Sharealike.
Additional terms may apply for the media files.