MYLIP
Myosin regulatory light chain interacting protein, also known as MYLIP, is a protein that in humans is encoded by the MYLIP gene.[5]
MYLIP is also known as IDOL "Inducible Degrader of the LDL receptor" based on its involvement in cholesterol regulation.[6][7] The expression of IDOL is induced by the sterol-activated liver X receptor.
Increased Degradation of LDL Receptor Protein (IDOL) is a ubiquitin ligase that ubiquinates LDL receptors in endosomes and directs the receptors to the lysosomal compartment for degradation. IDOL is transcriptionally up-regulated by LXR/RXR in response to an increase in intracellular cholesterol.[8] Pharmacologic inhibition of IDOL could reduce plasma LDL cholesterol by increasing plasma LDL receptor density.
Function
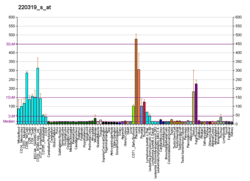
The ERM protein family members ezrin, radixin, and moesin are cytoskeletal effector proteins linking actin to membrane-bound proteins at the cell surface. Myosin regulatory light chain interacting protein (MYLIP) is a novel ERM-like protein that interacts with myosin regulatory light chain and inhibits neurite outgrowth.[5]
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000007944 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000038175 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- 1 2 "Entrez Gene: MYLIP myosin regulatory light chain interacting protein".
- ↑ Zelcer N, Hong C, Boyadjian R, Tontonoz P (July 2009). "LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor". Science. 325 (5936): 100–4. doi:10.1126/science.1168974. PMC 2777523. PMID 19520913.
- ↑ Lindholm D, Bornhauser BC, Korhonen L (November 2009). "Mylip makes an Idol turn into regulation of LDL receptor". Cell. Mol. Life Sci. 66 (21): 3399–402. doi:10.1007/s00018-009-0127-y. PMID 19688294.
- ↑ Sawamura, T. (2009). "New Idol for cholesterol reduction?". Clin. Chem. 55 (12): 2082–2084. doi:10.1373/clinchem.2009.134023.
Further reading
- Olsson PA, Korhonen L, Mercer EA, Lindholm D (2000). "MIR is a novel ERM-like protein that interacts with myosin regulatory light chain and inhibits neurite outgrowth". J. Biol. Chem. 274 (51): 36288–92. doi:10.1074/jbc.274.51.36288. PMID 10593918.
- Zhang QH, Ye M, Wu XY, et al. (2001). "Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells". Genome Res. 10 (10): 1546–60. doi:10.1101/gr.140200. PMC 310934. PMID 11042152.
- Olsson PA, Bornhauser BC, Korhonen L, Lindholm D (2001). "Neuronal expression of the ERM-like protein MIR in rat brain and its localization to human chromosome 6". Biochem. Biophys. Res. Commun. 279 (3): 879–83. doi:10.1006/bbrc.2000.4028. PMID 11162443.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Bornhauser BC, Olsson PA, Lindholm D (2003). "MSAP is a novel MIR-interacting protein that enhances neurite outgrowth and increases myosin regulatory light chain". J. Biol. Chem. 278 (37): 35412–20. doi:10.1074/jbc.M306271200. PMID 12826659.
- Bornhauser BC, Johansson C, Lindholm D (2003). "Functional activities and cellular localization of the ezrin, radixin, moesin (ERM) and RING zinc finger domains in MIR". FEBS Lett. 553 (1–2): 195–9. doi:10.1016/S0014-5793(03)01010-X. PMID 14550572.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Wan D, Gong Y, Qin W, et al. (2004). "Large-scale cDNA transfection screening for genes related to cancer development and progression". Proc. Natl. Acad. Sci. U.S.A. 101 (44): 15724–9. doi:10.1073/pnas.0404089101. PMC 524842. PMID 15498874.
- Rual JF, Venkatesan K, Hao T, et al. (2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. doi:10.1038/nature04209. PMID 16189514.
- Ohmura-Hoshino M, Matsuki Y, Aoki M, et al. (2006). "Inhibition of MHC class II expression and immune responses by c-MIR". J. Immunol. 177 (1): 341–54. doi:10.4049/jimmunol.177.1.341. PMID 16785530.