Plant lipid transfer proteins
| Plant lipid transfer protein | |||||||||
|---|---|---|---|---|---|---|---|---|---|
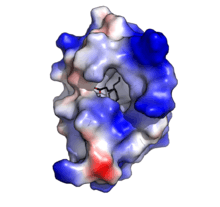 Oryza sativa Lipid Transfer Protein 1 bound to Palmitic acid (black). Positive charge in blue, negative charge in red. (PDB:1UVB ) | |||||||||
| Identifiers | |||||||||
| Symbol | LTP/seed_store/tryp_amyl_inhib | ||||||||
| Pfam | PF00234 | ||||||||
| InterPro | IPR003612 | ||||||||
| SMART | SM00499 | ||||||||
| |||||||||
Plant lipid transfer proteins, also known as plant LTPs or PLTPs, are a group of highly-conserved proteins of about 7-9kDa found in higher plant tissues.[1][2] As its name implies, lipid transfer proteins are responsible for the shuttling of phospholipids and other fatty acid groups between cell membranes.[3] LTPs are divided into two structurallyrelated subfamilies according to their molecular masses: LTP1s (9 kDa) and LTP2s (7 kDa).[4] Various LTPs bind a wide range of ligands, including fatty acids (FAs) with a C10–C18 chain length, acyl derivatives of coenzyme A (CoA), phospho- and galactolipids, prostaglandin B2, sterols, molecules of organic solvents, and some drugs.[2] LTP2
Biological activity
LTPs constitute one of the classes of defense PRPs, many of which have antimicrobial and enzymatic activities or are enzyme inhibitors. Different proteins of this class can exhibit the following activities:[2]
- antibacterial
- antifungal
- antiviral
- antiproliferative
and inhibit some enzymes.
Function
Ordinarily, most lipids do not spontaneously exit membranes because their hydrophobicity makes them poorly soluble in water. LTPs facilitate the movement of lipids between membranes by binding, and solubilising them. LTPs typically have broad substrate specificity and so can interact with a variety of different lipids.[5]
LTPs in plants may be involved in:
- cutin biosynthesis
- surface wax formation
- mitochondrial growth
- pathogen defense reactions
- adaptation to environmental changes[6]
- lipid metabolism
- fertilization of flowering plants
- adaptation of plants under stress conditions
- activation and regulation of signaling cascades
- apoptosis
- symbiosis
- fruit ripening[2]
Structure
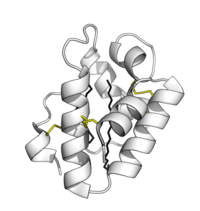

Plant lipid transfer proteins consist of 4 alpha-helices in a right-handed superhelix with a folded leaf topology. The structure is stabilised by disulfide bonds linking the helices to each other.
The structure forms an internal hydrophobic cavity in which 1-2 lipids can be bound. The outer surface of the protein is hydrophilic allowing the complex to be soluble. The use of hydrophobic interactions, with very few charged interactions, allows the protein to have broad specificity for a range of lipids.[5]
Other related proteins
Plant lipid transfer proteins share the same structural domain[7] with seed storage proteins[8] and trypsin-alpha amylase inhibitors.[9][10] These proteins share the same superhelical, disulphide-stabilised four-helix bundle containing an internal cavity.
There is no sequence similarity between animal and plant LTPs. In animals, cholesterylester transfer protein (CETP), also called plasma lipid transfer protein, is a plasma protein that facilitates the transport of cholesteryl esters and triglycerides between the lipoproteins.
Role in human health
PLTPs are pan-allergens, [11] [12] and may be directly responsible for cases of food allergy. Pru p 3, the major allergen from peach, is a 9-kDa allergen belonging to the family of lipid-transfer proteins. [13] Allergic properties are closely linked with high thermal stability and resistance to gastrointestinal proteolysis of the proteins.[14] Many of the LTP allergens are able to cause not only manifestation of allergic reactions but also sensitization via the gastrointestinal tract, beeng thus class I food allergens.[4] In contrast, class II food allergens are able only to elicit allergic symptoms due to its cross-reactivity with major sensitizer.
They are used as antioxidants and prevent diseases.[15]
Commercial importance
Lipid transfer protein 1 (from barley) is responsible, when denatured by the mashing process, for the bulk of foam which forms on top of beer.[16]
See also
References
- ↑ Asero R, Mistrello G, Roncarolo D, de Vries SC, Gautier MF, Ciurana CL, Verbeek E, Mohammadi T, Knul-Brettlova V, Akkerdaas JH, Bulder I, Aalberse RC, van Ree R (2001). "Lipid transfer protein: a pan-allergen in plant-derived foods that is highly resistant to pepsin digestion". International Archives of Allergy and Immunology. 124 (1–3): 67–9. doi:10.1159/000053671. PMID 11306929.
- 1 2 3 4 Finkina EI, Melnikova DN, Bogdanov IV, Ovchinnikova TV (2016). "Lipid Transfer Proteins As Components of the Plant Innate Immune System: Structure, Functions, and Applications". Acta Naturae. 8 (2): 47–61. PMC 4947988. PMID 27437139.
- ↑ Kader JC (June 1996). "Lipid-Transfer Protein in Plants". Annual Review of Plant Physiology and Plant Molecular Biology. 47: 627–654. doi:10.1146/annurev.arplant.47.1.627. PMID 15012303.
- 1 2 Finkina EI, Melnikova DN, Bogdanov IV, Ovchinnikova TV (2017-07-04). "Plant Pathogenesis-Related Proteins PR-10 and PR-14 as Components of Innate Immunity System and Ubiquitous Allergens". Current Medicinal Chemistry. 24 (17): 1772–1787. doi:10.2174/0929867323666161026154111. PMID 27784212.
- 1 2 Cheng HC, Cheng PT, Peng P, Lyu PC, Sun YJ (September 2004). "Lipid binding in rice nonspecific lipid transfer protein-1 complexes from Oryza sativa". Protein Science. 13 (9): 2304–15. doi:10.1110/ps.04799704. PMC 2280015. PMID 15295114.
- ↑ "Science Direct". Trends in Plant Science. 2: 66–70. doi:10.1016/S1360-1385(97)82565-4.
- ↑ Lin KF, Liu YN, Hsu ST, Samuel D, Cheng CS, Bonvin AM, Lyu PC (April 2005). "Characterization and structural analyses of nonspecific lipid transfer protein 1 from mung bean". Biochemistry. 44 (15): 5703–12. doi:10.1021/bi047608v. PMID 15823028.
- ↑ Pantoja-Uceda D, Bruix M, Giménez-Gallego G, Rico M, Santoro J (December 2003). "Solution structure of RicC3, a 2S albumin storage protein from Ricinus communis". Biochemistry. 42 (47): 13839–47. doi:10.1021/bi0352217. PMID 14636051.
- ↑ Oda Y, Matsunaga T, Fukuyama K, Miyazaki T, Morimoto T (November 1997). "Tertiary and quaternary structures of 0.19 alpha-amylase inhibitor from wheat kernel determined by X-ray analysis at 2.06 A resolution". Biochemistry. 36 (44): 13503–11. doi:10.1021/bi971307m. PMID 9354618.
- ↑ Gourinath S, Alam N, Srinivasan A, Betzel C, Singh TP (March 2000). "Structure of the bifunctional inhibitor of trypsin and alpha-amylase from ragi seeds at 2.2 A resolution". Acta Crystallographica D. 56 (Pt 3): 287–93. doi:10.1107/s0907444999016601. PMID 10713515.
- ↑ Morris A. "Food Allergy in Detail". Surrey Allergy Clinic.
- ↑ InterPro: IPR000528
- ↑ Besler M, Herranz JC, Fernández-Rivas M (2000). "Peach allergy". Internet Symposium on Food Allergens. 2 (4): 185–201.
- ↑ Bogdanov IV, Shenkarev ZO, Finkina EI, Melnikova DN, Rumynskiy EI, Arseniev AS, Ovchinnikova TV (April 2016). "A novel lipid transfer protein from the pea Pisum sativum: isolation, recombinant expression, solution structure, antifungal activity, lipid binding, and allergenic properties". BMC Plant Biology. 16: 107. doi:10.1186/s12870-016-0792-6. PMC 4852415. PMID 27137920.
- ↑ Halliwell B (1996). "Antioxidants in human health and disease". Annual Review of Nutrition. 16: 33–50. doi:10.1146/annurev.nu.16.070196.000341. PMID 8839918.
- ↑ "Foam". Carlsberg Research Laboratory.