LUC7L2
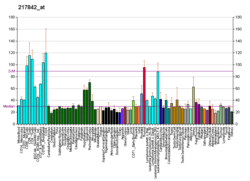
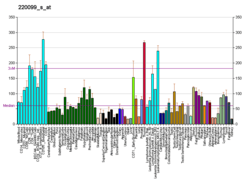
Putative RNA-binding protein Luc7-like 2 is a protein that in humans is encoded by the LUC7L2 gene.[5][6]
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000146963 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000029823 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Lai CH, Chou CY, Ch'ang LY, Liu CS, Lin W (Aug 2000). "Identification of Novel Human Genes Evolutionarily Conserved in Caenorhabditis elegans by Comparative Proteomics". Genome Res. 10 (5): 703–13. doi:10.1101/gr.10.5.703. PMC 310876. PMID 10810093.
- ↑ "Entrez Gene: LUC7L2 LUC7-like 2 (S. cerevisiae)".
Further reading
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Hillier LW, Fulton RS, Fulton LA, et al. (2003). "The DNA sequence of human chromosome 7". Nature. 424 (6945): 157–64. doi:10.1038/nature01782. PMID 12853948.
- Ota T, Suzuki Y, Nishikawa T, et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
- Colland F, Jacq X, Trouplin V, et al. (2004). "Functional Proteomics Mapping of a Human Signaling Pathway". Genome Res. 14 (7): 1324–32. doi:10.1101/gr.2334104. PMC 442148. PMID 15231748.
- Jin J, Smith FD, Stark C, et al. (2004). "Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization". Curr. Biol. 14 (16): 1436–50. doi:10.1016/j.cub.2004.07.051. PMID 15324660.
- Goehler H, Lalowski M, Stelzl U, et al. (2004). "A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease". Mol. Cell. 15 (6): 853–65. doi:10.1016/j.molcel.2004.09.016. PMID 15383276.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The Status, Quality, and Expansion of the NIH Full-Length cDNA Project: The Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Andersen JS, Lam YW, Leung AK, et al. (2005). "Nucleolar proteome dynamics". Nature. 433 (7021): 77–83. doi:10.1038/nature03207. PMID 15635413.
- Stelzl U, Worm U, Lalowski M, et al. (2005). "A human protein-protein interaction network: a resource for annotating the proteome". Cell. 122 (6): 957–68. doi:10.1016/j.cell.2005.08.029. PMID 16169070.
- Kimura K, Wakamatsu A, Suzuki Y, et al. (2006). "Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes". Genome Res. 16 (1): 55–65. doi:10.1101/gr.4039406. PMC 1356129. PMID 16344560.
- Olsen JV, Blagoev B, Gnad F, et al. (2006). "Global, in vivo, and site-specific phosphorylation dynamics in signaling networks". Cell. 127 (3): 635–48. doi:10.1016/j.cell.2006.09.026. PMID 17081983.
- Ewing RM, Chu P, Elisma F, et al. (2007). "Large-scale mapping of human protein–protein interactions by mass spectrometry". Mol. Syst. Biol. 3 (1): 89. doi:10.1038/msb4100134. PMC 1847948. PMID 17353931.
This article is issued from
Wikipedia.
The text is licensed under Creative Commons - Attribution - Sharealike.
Additional terms may apply for the media files.