Homoplasy
Homoplasy (derived from the Greek homo "the same" and plassein "to mold" [1][2]), in biological systematics, is when a trait has been gained or lost independently in separate lineages over the course of evolution. This is different from homology, which is the similarity of traits due to common ancestry. The term homoplasy was first used by R. Lankester in 1870 [3]. Homoplasy can arise from both similar selection pressures acting on adapting species, and the effects of genetic drift [4][5].
Parallel and convergent evolution lead to homoplasy when different species independently evolve or gain a comparable trait, which diverges from the trait inferred to have been present in their common ancestor. When the similar traits are caused by an equivalent developmental mechanism, the process is referred to as parallel evolution[6]. The process is called convergent evolution when the similarity arises from different developmental mechanisms [7]. These types of homoplasy may occur when different lineages live in comparable ecological niches that require similar adaptations for an increase in fitness. An interesting example is that of the marsupial moles (Notoryctidae), golden moles (Chrysochloridae) and northern moles (Talpidae). These are mammals from different geographical regions and lineages, and have all independently evolved very similar burrowing characteristics (such as a cone-shaped heads and flat frontal claws) to live in a subterranean ecological niche [8].
In contrast, reversal (a.k.a. vestigialization) leads to homoplasy through the disappearance of previously gained traits [9]. This process may result from changes in the environment in which certain gained traits are no longer relevant, or have even become costly [10][5]. This can be observed in subterranean and cave-dwelling animals by their loss of sight [8][11], in cave-dwelling animals through their loss of pigmentation [11], and in both snakes and legless lizards through their loss of limbs [12][13].
Most often, homoplasy is viewed as a similarity in morphological traits. However, homoplasy may also appear in other trait types, such as similarity in the genetic sequence [2][14], life cycle types [15] or even behavioral traits [16][14].
Distinguishing homology from homoplasy
Homoplasy, especially the type that occurs in more closely related phylogenetic groups, can make phylogenetic analysis more challenging. Phylogenetic trees are created by means of parsimony analysis [17][18]. These analyses can be done with phenotypic, as well as genetic traits (DNA sequences) [19]. Using parsimony analysis, the hypothesis that requires the fewest evolutionary changes is preferred over alternative hypotheses. Construction of these trees may become a challenge when clouded by the occurrence of homoplasy in the traits used for the analysis. The most important approach in overcoming these challenges, is by increasing the amount of independent (non-pleiotropic, non-linked) characteristics used in the construction of these phylogenic trees. Along with parsimony analysis, one could perform a likelihood analysis, where the probability of a tree being true is calculated and branch lengths are measured; and bootstrapping, in which trees are constructed for each characteristic separately to estimate the confidence of a tree [14].
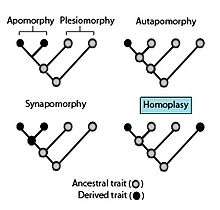
According to cladistic interpretation, homoplasy can be identified when a given similarity in trait cannot be explained by relation through a common ancestor on a preferred phylogenetic hypothesis - that is, the feature in question arises (or disappears) at more than one point on the tree [9].
In the case of DNA sequences, homoplasy cannot be avoided due to its four-state nature. An observed homoplasy may simply be the result of random nucleotide substitutions accumulating over time, and thus may not need an adaptationist evolutionary explanation [14].
Homoplasy vs. evolutionary contingency
In his book Wonderful Life, Stephen Jay Gould claims that repeating the evolutionary process, from any point in time onward, would not produce the same results [20]. The occurrence of homoplasy is viewed by some biologists as an argument against Gould’s theory of evolutionary contingency. Powell & Mariscal (2015) argue that this disagreement is caused by an equivocation and that both the theory of contingency as well as homoplastic occurrence can be true at the same time [21].
Application
The occurrence of homoplasy can also be used to make predictions about evolution. Recent studies have used homoplasy to predict the possibility and the path of extraterrestrial evolution. For example, Levin et al. (2017) suggest that the development of eye-like structures is highly likely, due to its numerous, independently evolved incidences on earth [9][22].
References
- ↑ Holt, J.R.; Judica, C.A. (February 4, 2014). "Systematic Biology - Dictionary of Terms: Homoplasy". Retrieved September 21, 2018.
- 1 2 Reece, J.B.; Urry, L.A.; Cain, M.L.; Wasserman, S.A.; Minorsky, P.V.; Jackson, R.B. (2011). Campbell Biology (9th ed.). Pearson. ISBN 9780321739759.
- ↑ Lankester, E.R. (1870). "On the use of the term homology in modern zoology, and the distinction between homogenetic and homoplastic agreements". Annals and Magazine of Natural History. 6: 34–43. doi:10.1080/00222937008696201.
- ↑ Stearns, S.C.; Hoekstra, R.F. (2005). Evolution: an introduction (2nd ed.). Oxford: Oxford University Press. ISBN 9780199255634.
- 1 2 Hall, A.R.; Colegrave, N. (2008). "Decay of unused characters by selection and drift". Journal of Evolutionary Biology. 21: 610–17. doi:10.1111/j.1420-9101.2007.01473.x.
- ↑ Archie, J.W. (1989). "Homoplasy excess ratios: new indices for measuring levels of homoplasy in phylogenetic systematics and a critique of the consistency index". Systematic Biology. 38: 253–269. doi:10.2307/2992286.
- ↑ Hodin, J. (2000). "Plasticity and constraints in development and evolution". Journal of Experimental Zoology. 288: 1–20. doi:10.1002/(SICI)1097-010X(20000415)288:1<1::AID-JEZ1>3.0.CO;2-7.
- 1 2 Nevo, E. (1979). "Adaptive convergence and divergence of subterranean mammals". Annual Review of Ecology and Systematics. 10: 269–308. doi:10.1146/annurev.es.10.110179.001413.
- 1 2 3 Wake, D.B.; Wake, M.H.; Specht, C.D. (2011). "Homoplasy: From detecting pattern to determining process and mechanism of evolution". Science. 331: 1032–1035. doi:10.1126/science.1188545.
- ↑ Fong, D.W.; Kane, T.C.; Culver, D.C. (1995). "Vestigialization and loss of nonfunctional characters". Annual Review of Ecology and Systematics. 26: 249–68. doi:10.1146/annurev.es.26.110195.001341.
- 1 2 Jones, R.; Culver, D.C. (1989). "Evidence for selection on sensory structures in a cave population of Gammarus minus (Amphipoda)". Evolution. 43: 688–693. doi:10.1111/j.1558-5646.1989.tb04267.x.
- ↑ Skinner, A.; Lee, M.S.Y. (2009). "Body-form evolution in the scincid lizard Lerista and the mode of macroevolutionary transitions". Evolutionary Biology. 36: 292–300. doi:10.1007/s11692-009-9064-9.
- ↑ Skinner, A; Lee, M.S.Y.; Hutchinson, M.N. (2008). "Rapid and repeated limb loss in a clade of scincid lizards". BMC Evolutionary Biology. 8: 310. doi:10.1186/1471-2148-8-310.
- 1 2 3 4 Sanderson, M.J.; Hufford, L. (1996). Homoplasy: The Recurrence of Similarity in Evolution. San Diego, CA: Academic Press, Inc. ISBN 0-12-618030-X.
- ↑ Silberfeld, T.; Leigh, J.W.; Verbruggen, H.; Cruaud, C.; de Reviers, B.; Rousseau, F. (2010). "A multi-locus time-calibrated phylogeny of the brown algae (Heterokonta, Ochrophyta, Phaeophyceae): Investigating the evolutionary nature of the "brown algal crown radiation."". Molecular Phylogenetics and Evolution. 56: 659–674. doi:10.1016/j.ympev.2010.04.020.
- ↑ de Querioz, A.; Wimberger, P.H. (1993). "The usefulness of behavior for phylogeny estimation: levels of homoplasy in behavioral and morphological characters". Evolution. 47: 46–60. doi:10.1111/j.1558-5646.1993.tb01198.x.
- ↑ Wiley, E.O.; Lieberman, B.S. (2011). Phylogenetics: Theory and Practice of Phylogenetic Systematics. Hoboken, NJ: John Wiley & Sons, Inc. ISBN 9780470905968.
- ↑ Schuh, R.T.; Brower, A.V.Z. (2018). Biological Systematics: Principles and Applications. Ithaca, NY: Cornell University Press. ISBN 9780801462436.
- ↑ Felsenstein, J. (2004). Inferring phylogenies. Sinauer. ISBN 978-0878931774.
- ↑ Gould, S.J. (2000). Wonderful Life: The Burgess Shale and the Nature of History. London: Vintage Books. ISBN 9780099273455.
- ↑ Powell, R.; Mariscal, C. (2015). "Convergent evolution as natural experiment: the tape of life reconsidered". Interface Focus. 5: 20150040. doi:10.1098/rsfs.2015.0040.
- ↑ Levin, S.R.; Scott, T.W.; Cooper, H.S.; West, S.A. (2017). "Darwin's aliens". International Journal of Astrobiology: 1–9. doi:10.1017/S1473550417000362.