HIST1H3B
Histone H3.1 is a protein that in humans is encoded by the HIST1H3B gene.[5][6][7]
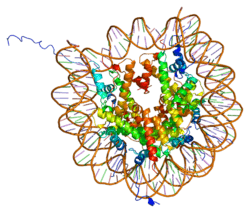
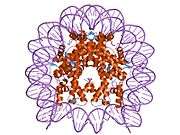
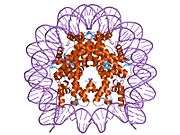
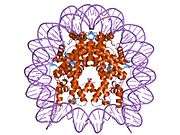
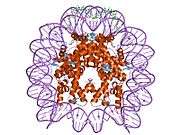
Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. This structure consists of approximately 146 bp of DNA wrapped around a nucleosome, an octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene is intronless and encodes a member of the histone H3 family. Transcripts from this gene lack polyA tails; instead, they contain a palindromic termination element. This gene is found in the large histone gene cluster on chromosome 6p22-p21.3.[8]
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000274267 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000074403 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Zhong R, Roeder RG, Heintz N (Jan 1984). "The primary structure and expression of four cloned human histone genes". Nucleic Acids Res. 11 (21): 7409–25. doi:10.1093/nar/11.21.7409. PMC 326492. PMID 6647026.
- ↑ Albig W, Kioschis P, Poustka A, Meergans K, Doenecke D (Apr 1997). "Human histone gene organization: nonregular arrangement within a large cluster". Genomics. 40 (2): 314–22. doi:10.1006/geno.1996.4592. PMID 9119399.
- ↑ Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ (Oct 2002). "The human and mouse replication-dependent histone genes". Genomics. 80 (5): 487–98. doi:10.1016/S0888-7543(02)96850-3. PMID 12408966.
- ↑ "Entrez Gene: HIST1H3B histone cluster 1, H3b".
Further reading
- Albig W, Kardalinou E, Drabent B, et al. (1991). "Isolation and characterization of two human H1 histone genes within clusters of core histone genes". Genomics. 10 (4): 940–8. doi:10.1016/0888-7543(91)90183-F. PMID 1916825.
- Marashi F, Helms S, Shiels A, et al. (1986). "Enhancer-facilitated expression of prokaryotic and eukaryotic genes using human histone gene 5' regulatory sequences". Biochem. Cell Biol. 64 (4): 277–89. doi:10.1139/o86-039. PMID 3013246.
- Ohe Y, Iwai K (1982). "Human spleen histone H3. Isolation and amino acid sequence". J. Biochem. 90 (4): 1205–11. PMID 7309716.
- Kardalinou E, Eick S, Albig W, Doenecke D (1993). "Association of a human H1 histone gene with an H2A pseudogene and genes encoding H2B.1 and H3.1 histones". J. Cell. Biochem. 52 (4): 375–83. doi:10.1002/jcb.240520402. PMID 8227173.
- Albig W, Meergans T, Doenecke D (1997). "Characterization of the H1.5 gene completes the set of human H1 subtype genes". Gene. 184 (2): 141–8. doi:10.1016/S0378-1119(96)00582-3. PMID 9031620.
- Albig W, Doenecke D (1998). "The human histone gene cluster at the D6S105 locus". Hum. Genet. 101 (3): 284–94. doi:10.1007/s004390050630. PMID 9439656.
- El Kharroubi A, Piras G, Zensen R, Martin MA (1998). "Transcriptional Activation of the Integrated Chromatin-Associated Human Immunodeficiency Virus Type 1 Promoter". Mol. Cell. Biol. 18 (5): 2535–44. doi:10.1128/mcb.18.5.2535. PMC 110633. PMID 9566873.
- Goto H, Tomono Y, Ajiro K, et al. (1999). "Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation". J. Biol. Chem. 274 (36): 25543–9. doi:10.1074/jbc.274.36.25543. PMID 10464286.
- Hsu JY, Sun ZW, Li X, et al. (2000). "Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes". Cell. 102 (3): 279–91. doi:10.1016/S0092-8674(00)00034-9. PMID 10975519.
- Deng L, de la Fuente C, Fu P, et al. (2001). "Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones". Virology. 277 (2): 278–95. doi:10.1006/viro.2000.0593. PMID 11080476.
- Lachner M, O'Carroll D, Rea S, et al. (2001). "Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins". Nature. 410 (6824): 116–20. doi:10.1038/35065132. PMID 11242053.
- Deng L, Wang D, de la Fuente C, et al. (2001). "Enhancement of the p300 HAT activity by HIV-1 Tat on chromatin DNA". Virology. 289 (2): 312–26. doi:10.1006/viro.2001.1129. PMID 11689053.
- Yang L, Xia L, Wu DY, et al. (2002). "Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor". Oncogene. 21 (1): 148–52. doi:10.1038/sj.onc.1204998. PMID 11791185.
- Goto H, Yasui Y, Nigg EA, Inagaki M (2002). "Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation". Genes Cells. 7 (1): 11–7. doi:10.1046/j.1356-9597.2001.00498.x. PMID 11856369.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Preuss U, Landsberg G, Scheidtmann KH (2003). "Novel mitosis-specific phosphorylation of histone H3 at Thr11 mediated by Dlk/ZIP kinase". Nucleic Acids Res. 31 (3): 878–85. doi:10.1093/nar/gkg176. PMC 149197. PMID 12560483.
- Mungall AJ, Palmer SA, Sims SK, et al. (2003). "The DNA sequence and analysis of human chromosome 6". Nature. 425 (6960): 805–11. doi:10.1038/nature02055. PMID 14574404.