H2AFZ
Histone H2A.Z is a protein that in humans is encoded by the H2AFZ gene.[5][6]
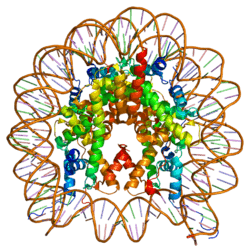
Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Nucleosomes consist of approximately 146 bp of DNA wrapped around a histone octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. The H2AFZ gene encodes a replication-independent member of the histone H2A family that is distinct from other members of the family. Studies in mice have shown that this particular histone is required for embryonic development and indicate that lack of functional histone H2A leads to embryonic lethality.[6]
Histone H2AZ is a variant of histone H2A, and is used to mediate the thermosensory response, and is essential to perceive the ambient temperature. Nucleosome occupancy of H2A.Z decreases with temperature, and in vitro assays show that H2A.Z-containing nucleosomes wrap DNA more tightly than canonical H2A nucleosomes in Arabidopsis.(Cell 140: 136–147, 2010) However, in some of the other studies (Nat. Genet. 41, 941–945 and Genes Dev., 21, 1519–1529) have shown that incorporation of H2A.Z in nucleosomes, when it co-occurs with H3.3, makes them weaker. Positioning of H2A.Z containing nucleosomes around transcription start sites has now been shown to affect the downstream gene expression.[7]
Recent evidence also points to a role for H2A.Z in repressing a subset of ncRNAs, derepressing CUTs, as well as mediation higher order chromatin structure formation.[8]
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000164032 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000037894 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Hatch CL, Bonner WM (Oct 1990). "The human histone H2A.Z gene. Sequence and regulation". J Biol Chem. 265 (25): 15211–8. PMID 1697587.
- 1 2 "Entrez Gene: H2AFZ H2A histone family, member Z".
- ↑ Bargaje R., Alam P., Patowary A., Sarkar M., Ali T., Gupta S., Garg M., Singh M., Purkanti R., Scaria V., Sivasubbu S., Brahmachari V., Pillai B. (2012) Proximity of H2A.Z containing nucleosome to the transcription start site influences gene expression levels in the mammalian liver and brain. Nucleic Acids Research (Epub ahead of print)Bargaje R, Alam MP, Patowary A, et al. (October 2012). "Proximity of H2A.Z containing nucleosome to the transcription start site influences gene expression levels in the mammalian liver and brain". Nucleic Acids Research. 40 (18): 8965–78. doi:10.1093/nar/gks665. PMC 3467062. PMID 22821566.
- ↑ Mayuri Rege, Vidya Subramanian, Chenchen Zhu, Tsung-Han S. Hsieh, Assaf Weiner,Nir Friedman,Sandra Clauder-Mu€nster, Lars M. Steinmetz, Oliver J. Rando,Laurie A. Boyer, and Craig L. Peterson. (2015) Chromatin Dynamics and the RNA Exosome Function in Concert to Regulate Transcriptional Homeostasis doi:10.1016/j.celrep.2015.10.030
Further reading
- Jason LJ, Moore SC, Lewis JD, et al. (2002). "Histone ubiquitination: a tagging tail unfolds?". BioEssays. 24 (2): 166–74. doi:10.1002/bies.10038. PMID 11835281.
- Hatch CL, Bonner WM (1988). "Sequence of cDNAs for mammalian H2A.Z, an evolutionarily diverged but highly conserved basal histone H2A isoprotein species". Nucleic Acids Res. 16 (3): 1113–24. doi:10.1093/nar/16.3.1113. PMC 334740. PMID 3344202.
- Kato S, Sekine S, Oh SW, et al. (1995). "Construction of a human full-length cDNA bank". Gene. 150 (2): 243–50. doi:10.1016/0378-1119(94)90433-2. PMID 7821789.
- Hatch CL, Bonner WM (1995). "Characterization of the proximal promoter of the human histone H2A.Z gene". DNA Cell Biol. 14 (3): 257–66. doi:10.1089/dna.1995.14.257. PMID 7880446.
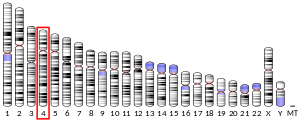
- Popescu N, Zimonjic D, Hatch C, Bonner W (1994). "Chromosomal mapping of the human histone gene H2AZ to 4q24 by fluorescence in situ hybridization". Genomics. 20 (2): 333–5. doi:10.1006/geno.1994.1182. PMID 8020992.
- Hatch CL, Bonner WM (1996). "An upstream region of the H2AZ gene promoter modulates promoter activity in different cell types". Biochim. Biophys. Acta. 1305 (1–2): 59–62. doi:10.1016/0167-4781(95)00223-5. PMID 8605251.
- El Kharroubi A, Piras G, Zensen R, Martin MA (1998). "Transcriptional Activation of the Integrated Chromatin-Associated Human Immunodeficiency Virus Type 1 Promoter". Mol. Cell. Biol. 18 (5): 2535–44. doi:10.1128/mcb.18.5.2535. PMC 110633. PMID 9566873.
- Slachta CA, Jeevanandam V, Goldman B, et al. (2000). "Coronary arteries from human cardiac allografts with chronic rejection contain oligoclonal T cells: persistence of identical clonally expanded TCR transcripts from the early post-transplantation period (endomyocardial biopsies) to chronic rejection (coronary arteries)". J. Immunol. 165 (6): 3469–83. doi:10.4049/jimmunol.165.6.3469. PMID 10975868.
- Pasqualucci L, Neri A, Baldini L, et al. (2000). "BCL-6 mutations are associated with immunoglobulin variable heavy chain mutations in B-cell chronic lymphocytic leukemia". Cancer Res. 60 (20): 5644–8. PMID 11059755.
- Deng L, de la Fuente C, Fu P, et al. (2001). "Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones". Virology. 277 (2): 278–95. doi:10.1006/viro.2000.0593. PMID 11080476.
- Suto RK, Clarkson MJ, Tremethick DJ, Luger K (2001). "Crystal structure of a nucleosome core particle containing the variant histone H2A.Z". Nat. Struct. Biol. 7 (12): 1121–4. doi:10.1038/81971. PMID 11101893.
- Bräuninger A, Yang W, Wacker HH, et al. (2001). "B-cell development in progressively transformed germinal centers: similarities and differences compared with classical germinal centers and lymphocyte-predominant Hodgkin disease". Blood. 97 (3): 714–9. doi:10.1182/blood.V97.3.714. PMID 11157489.
- Yamamoto K, Sugita N, Kobayashi T, et al. (2001). "Evidence for a novel polymorphism affecting both N-linked glycosylation and ligand binding of the IgG receptor IIIB (CD16)". Tissue Antigens. 57 (4): 363–6. doi:10.1034/j.1399-0039.2001.057004363.x. PMID 11380948.
- Faast R, Thonglairoam V, Schulz TC, et al. (2001). "Histone variant H2A.Z is required for early mammalian development". Curr. Biol. 11 (15): 1183–7. doi:10.1016/S0960-9822(01)00329-3. PMID 11516949.
- Deng L, Wang D, de la Fuente C, et al. (2001). "Enhancement of the p300 HAT activity by HIV-1 Tat on chromatin DNA". Virology. 289 (2): 312–26. doi:10.1006/viro.2001.1129. PMID 11689053.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Rangasamy D, Berven L, Ridgway P, Tremethick DJ (2003). "Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development". EMBO J. 22 (7): 1599–607. doi:10.1093/emboj/cdg160. PMC 152904. PMID 12660166.
- Lusic M, Marcello A, Cereseto A, Giacca M (2004). "Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter". EMBO J. 22 (24): 6550–61. doi:10.1093/emboj/cdg631. PMC 291826. PMID 14657027.