Histone H2A.V is a protein that in humans is encoded by the H2AFV gene.[5]
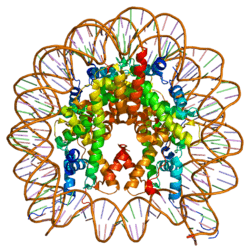
Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Nucleosomes consist of approximately 146 bp of DNA wrapped around a histone octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene encodes a member of the histone H2A family. Several transcripts have been identified for this gene.[5]
Further reading
- El Kharroubi A, Piras G, Zensen R, Martin MA (1998). "Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter". Mol. Cell. Biol. 18 (5): 2535–44. doi:10.1128/mcb.18.5.2535. PMC 110633. PMID 9566873.
- Deng L, de la Fuente C, Fu P, et al. (2001). "Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones". Virology. 277 (2): 278–295. doi:10.1006/viro.2000.0593. PMID 11080476.
- Deng L, Wang D, de la Fuente C, et al. (2001). "Enhancement of the p300 HAT activity by HIV-1 Tat on chromatin DNA". Virology. 289 (2): 312–326. doi:10.1006/viro.2001.1129. PMID 11689053.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–16903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Hillier LW, Fulton RS, Fulton LA, et al. (2003). "The DNA sequence of human chromosome 7". Nature. 424 (6945): 157–164. doi:10.1038/nature01782. PMID 12853948.
- Lusic M, Marcello A, Cereseto A, Giacca M (2004). "Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter". EMBO J. 22 (24): 6550–6561. doi:10.1093/emboj/cdg631. PMC 291826. PMID 14657027.
- Ota T, Suzuki Y, Nishikawa T, et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs". Nat. Genet. 36 (1): 40–45. doi:10.1038/ng1285. PMID 14702039.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–2127. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Oh JH, Yang JO, Hahn Y, et al. (2006). "Transcriptome analysis of human gastric cancer". Mamm. Genome. 16 (12): 942–954. doi:10.1007/s00335-005-0075-2. PMID 16341674.
- Kimura K, Wakamatsu A, Suzuki Y, et al. (2006). "Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes". Genome Res. 16 (1): 55–65. doi:10.1101/gr.4039406. PMC 1356129. PMID 16344560.
- Boyne MT, Pesavento JJ, Mizzen CA, Kelleher NL (2006). "Precise characterization of human histones in the H2A gene family by top down mass spectrometry". J. Proteome Res. 5 (2): 248–253. doi:10.1021/pr050269n. PMID 16457589.
- Kim SC, Sprung R, Chen Y, et al. (2006). "Substrate and functional diversity of lysine acetylation revealed by a proteomics survey". Mol. Cell. 23 (4): 607–618. doi:10.1016/j.molcel.2006.06.026. PMID 16916647.
PDB gallery |
|---|
1f66: 2.6 A CRYSTAL STRUCTURE OF A NUCLEOSOME CORE PARTICLE CONTAINING THE VARIANT HISTONE H2A.Z |