Gliding motility
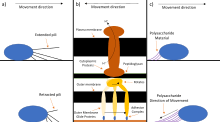
Gliding motility is a type of flagella-independent translocation that allows the microorganism to glide smoothly along a surface without the aid of propulsive organelles on the outer membrane.[6] Gliding motility and twitching motility both allow microorganisms to travel along the surface of low aqueous films, but while gliding motility is smooth, twitching motility is jerky and uses the pili as its mechanism of transport. Motor proteins found within the inner membrane of the bacteria utilize a proton-conducting channel to transduce a mechanical force to the cell surface.[6] The movement of the cytoskeletal filaments causes a mechanical force which travels to the adhesion complexes on the substrate to move the cell forward.[7] Motor and regulatory proteins that convert intracellular motion into mechanical forces like traction force have been discovered to be a conserved class of intracellular motors in bacteria that have been adapted to produce cell motility.[7] The speed of the gliding varies from organism to organism and the reversal of direction is seemingly regulated by an internal clock of some sort.[8] Gliding motility known as bacterial gliding in bacteria, is not unique to bacteria since it can also be seen being utilized by the Apicomplexa, a Eukaryota parasite, travelling at fast rates between 1–10 μm a second when the Myxococcus xanthus glide at a rate of 5 μm a minute.[9] Cell invasion and gliding motility have TRAP(thrombospondin-related anonymous protein), a surface protein, as a common molecular basis that is both essential for infection and locomotion of the invasive Apicomplexa parasite.[10]
See also
References
- ↑ Strom, M S; Lory, S (1993-10-01). "Structure-Function and Biogenesis of the Type Iv Pili". Annual Review of Microbiology. 47 (1): 565–596. doi:10.1146/annurev.mi.47.100193.003025. ISSN 0066-4227.
- ↑ McBride, Mark J. (2001-10-01). "Bacterial Gliding Motility: Multiple Mechanisms for Cell Movement over Surfaces". Annual Review of Microbiology. 55 (1): 49–75. doi:10.1146/annurev.micro.55.1.49. ISSN 0066-4227.
- ↑ Dzink-Fox, J. L.; Leadbetter, E. R.; Godchaux, W. (December 1997). "Acetate acts as a protonophore and differentially affects bead movement and cell migration of the gliding bacterium Cytophaga johnsonae (Flavobacterium johnsoniae)". Microbiology. 143 (12): 3693–3701. doi:10.1099/00221287-143-12-3693. ISSN 1350-0872. PMID 9421895.
- ↑ Braun, Timothy F.; Khubbar, Manjeet K.; Saffarini, Daad A.; McBride, Mark J. (September 2005). "Flavobacterium johnsoniae Gliding Motility Genes Identified by mariner Mutagenesis". Journal of Bacteriology. 187 (20): 6943–6952. doi:10.1128/JB.187.20.6943-6952.2005. ISSN 0021-9193. PMC 1251627. PMID 16199564.
- ↑ Hoiczyk, E.; Baumeister, W. (1998-10-22). "The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria". Current Biology. 8 (21): 1161–1168. doi:10.1016/s0960-9822(07)00487-3. ISSN 0960-9822. PMID 9799733.
- 1 2 Nan, Beiyan (February 2017). "Bacterial Gliding Motility: Rolling Out a Consensus Model". Current Biology. 27 (4): R154–R156. doi:10.1016/j.cub.2016.12.035.
- 1 2 Sun, Mingzhai; Wartel, Morgane; Cascales, Eric; Shaevitz, Joshua W.; Mignot, Tâm (2011-05-03). "Motor-driven intracellular transport powers bacterial gliding motility". Proceedings of the National Academy of Sciences. 108 (18): 7559–7564. doi:10.1073/pnas.1101101108. ISSN 0027-8424. PMC 3088616. PMID 21482768.
- ↑ Nan, Beiyan; McBride, Mark J.; Chen, Jing; Zusman, David R.; Oster, George (February 2014). "Bacteria that Glide with Helical Tracks". Current Biology. 24 (4): 169–174. doi:10.1016/j.cub.2013.12.034.
- ↑ Sibley, L. David; Håkansson, Sebastian; Carruthers, Vern B (1998-01-01). "Gliding motility: An efficient mechanism for cell penetration". Current Biology. 8 (1): R12–R14. doi:10.1016/S0960-9822(98)70008-9.
- ↑ Sultan, Ali A; Thathy, Vandana; Frevert, Ute; Robson, Kathryn J.H; Crisanti, Andrea; Nussenzweig, Victor; Nussenzweig, Ruth S; Ménard, Robert (1997). "TRAP Is Necessary for Gliding Motility and Infectivity of Plasmodium Sporozoites". Cell. 90 (3): 511–522. doi:10.1016/s0092-8674(00)80511-5.