GLYAT
Glycine-N-acyltransferase, also known as GLYAT, is an enzyme which in humans is encoded by the GLYAT gene.[5][6]
Function
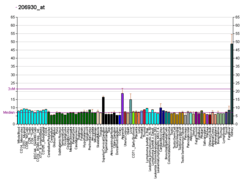
The glycine-N-acyltransferase protein conjugates glycine with acyl-CoA substrates in the mitochondria primarily in liver and kidney. The glycine N-acyltransferase enzyme is involved in the detoxification of a wide range of xenobiotic and endogenous metabolites. These include benzoic acid, a compound found in fruits and vegetables and used in medicine and foodstuffs as a preservative; salicylic acid, a metabolite of aspirin; and several endogenous metabolites. The diversity is demonstrated by the wide range of acylglycines excreted in the urines of patients with defects of organic acid metabolism. No defect of glycine N-acyltransferase has yet been described, but it has been demonstrated that there is significant inter individual variation in glycine conjugation capacity. Human glycine N-acyltransferase isoform a is a 296 amino acid protein translated from mRNA transcript splice variant 1. It is encoded by exons 2 to 6 of the mRNA transcript.[5]
Molecular weight
The literature reports it to be approximately 30 kDa,[6] or approximately 27 kDa.[7] The predicted size is 33.9 KDa. For the bovine enzyme a range of sizes between approximately 33 kDa and about 36 KDa is reported (Nandi, 1979, Vessey, 1992, van der Westhuizen, 2000). The predicted size of bovine GLYAT based on its sequence (accession number nm: 177486), is 33.9 kDa. This compares well to the experimentally determined sizes[7][8][9]
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000149124 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000063683 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- 1 2 "Entrez Gene: GLYAT glycine-N-acyltransferase".
- 1 2 Mawal YR, Qureshi IA (December 1994). "Purification to homogeneity of mitochondrial acyl coa:glycine n-acyltransferase from human liver". Biochem. Biophys. Res. Commun. 205 (2): 1373–9. doi:10.1006/bbrc.1994.2817. PMID 7802672.
- 1 2 van der Westhuizen FH, Pretorius PJ, Erasmus E (2000). "The utilization of alanine, glutamic acid, and serine as amino acid substrates for glycine N-acyltransferase". J. Biochem. Mol. Toxicol. 14 (2): 102–9. doi:10.1002/(SICI)1099-0461(2000)14:2<102::AID-JBT6>3.0.CO;2-H. PMID 10630424.
- ↑ Nandi DL, Lucas SV, Webster LT (August 1979). "Benzoyl-coenzyme A:glycine N-acyltransferase and phenylacetyl-coenzyme A:glycine N-acyltransferase from bovine liver mitochondria. Purification and characterization". J. Biol. Chem. 254 (15): 7230–7. PMID 457678.
- ↑ Kelley M, Vessey DA (November 1992). "Structural comparison between the mitochondrial aralkyl-CoA and arylacetyl-CoA N-acyltransferases". Biochem. J. 288 (1): 315–7. PMC 1132116. PMID 1445276.
Further reading
- Webster LT, Siddiqui UA, Lucas SV, et al. (1976). "Identification of separate acyl- CoA:glycine and acyl-CoA:L-glutamine N-acyltransferase activities in mitochondrial fractions from liver of rhesus monkey and man". J. Biol. Chem. 251 (11): 3352–8. PMID 931988.
- Mawal YR, Qureshi IA (1994). "Purification to homogeneity of mitochondrial acyl coa:glycine n-acyltransferase from human liver". Biochem. Biophys. Res. Commun. 205 (2): 1373–9. doi:10.1006/bbrc.1994.2817. PMID 7802672.
- Mawal YR, Qureshi IA (1995). "An immunodetection method for the quantitation of human acyl CoA:glycine N-acyltransferase in biological samples". Biochem. Mol. Biol. Int. 34 (3): 595–601. PMID 7833837.
- Merkler DJ, Merkler KA, Stern W, Fleming FF (1996). "Fatty acid amide biosynthesis: a possible new role for peptidylglycine alpha-amidating enzyme and acyl-coenzyme A: glycine N-acyltransferase". Arch. Biochem. Biophys. 330 (2): 430–4. doi:10.1006/abbi.1996.0272. PMID 8660675.
- Mawal Y, Paradis K, Qureshi IA (1997). "Developmental profile of mitochondrial glycine N-acyltransferase in human liver". J. Pediatr. 130 (6): 1003–7. doi:10.1016/S0022-3476(97)70293-2. PMID 9202629.
- van der Westhuizen FH, Pretorius PJ, Erasmus E (2000). "The utilization of alanine, glutamic acid, and serine as amino acid substrates for glycine N-acyltransferase". J. Biochem. Mol. Toxicol. 14 (2): 102–9. doi:10.1002/(SICI)1099-0461(2000)14:2<102::AID-JBT6>3.0.CO;2-H. PMID 10630424.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- SCHACHTER D, TAGGART JV (2003). "Glycine N-acylase: purification and properties". J. Biol. Chem. 208 (1): 263–75. PMID 13174534.
- Suzuki Y, Yamashita R, Shirota M, et al. (2004). "Sequence comparison of human and mouse genes reveals a homologous block structure in the promoter regions". Genome Res. 14 (9): 1711–8. doi:10.1101/gr.2435604. PMC 515316. PMID 15342556.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
External links
- GLYAT human gene location in the UCSC Genome Browser.
- GLYAT human gene details in the UCSC Genome Browser.