Peptide bond
A peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another along a peptide or protein chain.[1]
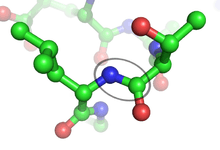
It can also be called an eupeptide bond[1] to separate it from an isopeptide bond, a different type of amide bond between two amino acids.
Synthesis
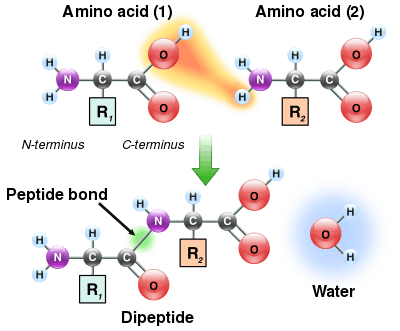
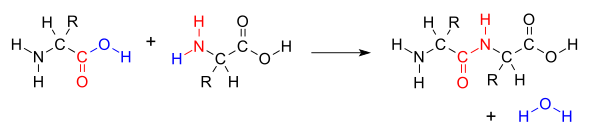
When two amino acids form a dipeptide through a peptide bond[1] it is a type of condensation reaction.[2] In this kind of condensation, two amino acids approach each other, with the non-side chain (C1) carboxylic acid moiety of one coming near the non-side chain (N2) amino moiety of the other. One loses a hydrogen and oxygen from its carboxyl group (COOH) and the other loses a hydrogen from its amino group (NH2). This reaction produces a molecule of water (H2O) and two amino acids joined by a peptide bond (-CO-NH-). The two joined amino acids are called a dipeptide.
The amide bond is synthesized when the carboxyl group of one amino acid molecule reacts with the amino group of the other amino acid molecule, causing the release of a molecule of water (H2O), hence the process is a dehydration synthesis reaction.
The formation of the peptide bond consumes energy, which, in organisms, is derived from ATP.[3] Peptides and proteins are chains of amino acids held together by peptide bonds (and sometimes by a few isopeptide bonds). Organisms use enzymes to produce nonribosomal peptides,[4] and ribosomes to produce proteins via reactions that differ in details from dehydration synthesis.[5]
Some peptides, like alpha-amanitin, are called ribosomal peptides as they are made by ribosomes,[6] but many are nonribosomal peptides as they are synthesized by specialized enzymes rather than ribosomes. For example, the tripeptide glutathione is synthesized in two steps from free amino acids, by two enzymes: glutamate–cysteine ligase (forms an isopeptide bond, which is not a peptide bond) and glutathione synthetase (forms a peptide bond).[7][8]
Degradation
A peptide bond can be broken by hydrolysis (the addition of water). In the presence of water they will break down and release 8–16 kilojoule/mol (2–4 kcal/mol) of Gibbs energy.[9] This process is extremely slow, with the half life at 25 °C of between 350 and 600 years per bond.[10]
In living organisms, the process is normally catalyzed by enzymes known as peptidases or proteases, although there are reports of peptide bond hydrolysis caused by conformational strain as the peptide/protein folds into the native structure.[11] This non-enzymatic process is thus not accelerated by transition state stabilization, but rather by ground state destabilization.
Spectra
The wavelength of absorption A for a peptide bond is 190–230 nm[12] (which makes it particularly susceptible to UV radiation).
Cis/trans isomers of the peptide group
Significant delocalisation of the lone pair of electrons on the nitrogen atom gives the group a partial double bond character. The partial double bond renders the amide group planar, occurring in either the cis or trans isomers. In the unfolded state of proteins, the peptide groups are free to isomerize and adopt both isomers; however, in the folded state, only a single isomer is adopted at each position (with rare exceptions). The trans form is preferred overwhelmingly in most peptide bonds (roughly 1000:1 ratio in trans:cis populations). However, X-Pro peptide groups tend to have a roughly 30:1 ratio, presumably because the symmetry between the and atoms of proline makes the cis and trans isomers nearly equal in energy (see figure, below).
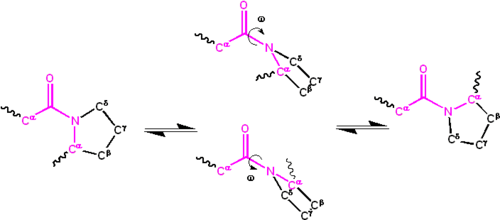
The dihedral angle associated with the peptide group (defined by the four atoms ) is denoted ; for the cis isomer (synperiplanar conformation) and for the trans isomer (antiperiplanar conformation). Amide groups can isomerize about the C'-N bond between the cis and trans forms, albeit slowly (20 seconds at room temperature). The transition states requires that the partial double bond be broken, so that the activation energy is roughly 80 kilojoule/mol (20 kcal/mol). However, the activation energy can be lowered (and the isomerization catalyzed) by changes that favor the single-bonded form, such as placing the peptide group in a hydrophobic environment or donating a hydrogen bond to the nitrogen atom of an X-Pro peptide group. Both of these mechanisms for lowering the activation energy have been observed in peptidyl prolyl isomerases (PPIases), which are naturally occurring enzymes that catalyze the cis-trans isomerization of X-Pro peptide bonds.
Conformational protein folding is usually much faster (typically 10–100 ms) than cis-trans isomerization (10–100 s). A nonnative isomer of some peptide groups can disrupt the conformational folding significantly, either slowing it or preventing it from even occurring until the native isomer is reached. However, not all peptide groups have the same effect on folding; nonnative isomers of other peptide groups may not affect folding at all.
Chemical reactions
Due to its resonance stabilization, the peptide bond is relatively unreactive under physiological conditions, even less than similar compounds such as esters. Nevertheless, peptide bonds can undergo chemical reactions, usually through an attack of an electronegative atom on the carbonyl carbon, breaking the carbonyl double bond and forming a tetrahedral intermediate. This is the pathway followed in proteolysis and, more generally, in N-O acyl exchange reactions such as those of inteins. When the functional group attacking the peptide bond is a thiol, hydroxyl or amine, the resulting molecule may be called a cyclol or, more specifically, a thiacyclol, an oxacyclol or an azacyclol, respectively.
See also
- The Proteolysis Map
References
- "Nomenclature and Symbolism for Amino Acids and Peptides. Recommendations 1983". European Journal of Biochemistry. 138 (1): 9–37. 1984. doi:10.1111/j.1432-1033.1984.tb07877.x. ISSN 0014-2956. PMID 6692818.
- Muller, P (1994-01-01). "Glossary of terms used in physical organic chemistry (IUPAC Recommendations 1994)". Pure and Applied Chemistry. 66 (5): 1077–1184. doi:10.1351/pac199466051077. ISSN 1365-3075.
- Watson J, Hopkins N, Roberts J, Agetsinger Steitz J, Weiner A (1987) [1965]. Molecualar Biology of the Gene (hardcover) (Fourth ed.). Menlo Park, CA: The Benjamin/Cummings Publishing Company, Inc. p. 168. ISBN 978-0805396140.
- Miller BR, Gulick AM (2016). "Structural Biology of Nonribosomal Peptide Synthetases". Methods in Molecular Biology. 1401: 3–29. doi:10.1007/978-1-4939-3375-4_1. ISBN 978-1-4939-3373-0. PMC 4760355. PMID 26831698.
- Griffiths AJ, Miller JH, Suzuki DT, Lewontin RC, Gelbart WM (2000). Protein synthesis. An Introduction to Genetic Analysis (7th ed.). New York: W. H. Freeman. ISBN 978-0716735205.
- Walton JD, Hallen-Adams HE, Luo H (2010). "Ribosomal biosynthesis of the cyclic peptide toxins of Amanita mushrooms". Biopolymers. 94 (5): 659–64. doi:10.1002/bip.21416. PMC 4001729. PMID 20564017.
- Wu G, Fang YZ, Yang S, Lupton JR, Turner ND (March 2004). "Glutathione metabolism and its implications for health". The Journal of Nutrition. 134 (3): 489–92. doi:10.1093/jn/134.3.489. PMID 14988435.
- Meister A (November 1988). "Glutathione metabolism and its selective modification". The Journal of Biological Chemistry. 263 (33): 17205–8. PMID 3053703.
- Martin RB (December 1998). "Free energies and equilibria of peptide bond hydrolysis and formation". Biopolymers. 45 (5): 351–353. doi:10.1002/(SICI)1097-0282(19980415)45:5<351::AID-BIP3>3.0.CO;2-K.
- Radzicka A, Wolfenden R (1996-01-01). "Rates of Uncatalyzed Peptide Bond Hydrolysis in Neutral Solution and the Transition State Affinities of Proteases". Journal of the American Chemical Society. 118 (26): 6105–6109. doi:10.1021/ja954077c. ISSN 0002-7863.
- Sandberg A, Johansson DG, Macao B, Härd T (April 2008). "SEA domain autoproteolysis accelerated by conformational strain: energetic aspects". Journal of Molecular Biology. 377 (4): 1117–29. doi:10.1016/j.jmb.2008.01.051. PMID 18308334.
- Goldfarb AR, Saidel LJ, Mosovich E (November 1951). "The ultraviolet absorption spectra of proteins". The Journal of Biological Chemistry. 193 (1): 397–404. PMID 14907727.