Ubiquitin bacterial
Ubiquitin Bacterial (UBact) is a ubiquitin-like protein that is homologus to Prokaryotic ubiquitin-like protein (Pup). UBact was recently described by the group of Professor Aaron Ciechanover at the Technion, Israel.[1]
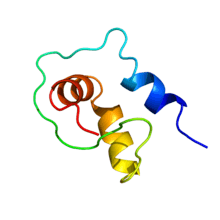
Ubiquitin was named for its ubiquitous presence among eukaryotes, while UBact ('Ubiquitin bacterial') is very limited in occurrence among the vast number of bacterial species.[1] The terms 'Ubiquitin Bacterial' and 'Prokaryotic ubiquitin-like protein' suggest a molecular similarity between ubiquitin and UBact/Pup which is largely absent.[2] While ubiquitin assumes a highly stable three-dimensional structure in solution,[3] Pup has been shown to belong to the group of intrinsically disordered proteins.[4][5]
The establishment of the term UBact is controversial, since to date there is no experimental evidence presented to justify the distinction of UBact from Pup.[1] The term UBact was denoted because several bacterial species from the phylum Nitrospirae (whrere UBact was initially identified; e.g., Leptospirillum ferriphilum) contain both the Pup-proteasome system[6] and a novel ORF-proteasome system that needed to be addressed[7] and therefore was denoted UBact.[1] The conjugation-proteasome components neighboring the UBact and Pup loci in these Nitrospirae bacteria show weak similarity and are probably not entirely redundant. Figure 2 illustrates the differences between the UBact and Pup loci in the representative Nitrospirae bacterium Leptospirillum ferrodiazotrophum. Further analyses of the UBact (and not Pup) locus in Leptospirillum ferrodiazotrophum revealed its existence and extreme conservation across several gram-negative bacterial phyla, as illustrated in figure 3.

In spite of the large difference in sequence, UBact is homologous to Pup and shares several characteristics with it: (i) same genomic location within a cluster of genes homologous to Mpa -> Dop -> Pup/UBact -> PrcB -> PrcA -> PafA, (ii) C-terminal sequence that ends exclusively with glutamine or glutamate across bacterial species, (iii) short size (similar to that of ubiquitin) and, (iv) high sequence conservation across tremendous evolutionary distance (a characteristic also in common with ubiquitin). The differences between UBact and Pup are their taxonomic distribution and amino acid sequences. While Pup is predominantly found in the gram-positive phylum Actinobacteria, UBact was identified only in gram-negative bacteria from the following five phyla: Nitrospirae, Verrucomicrobia, Armatimonadetes, Nitrospinae, and Planctomycetes. UBact was also identified in the genomes of several candidatus bacteria, and specifically from the candidate divisions Acetothermia, Handelsmanbacteria, Fraserbacteria, Terrybacteria, Poribacteria, Parcubacteria and Yanofskybacteria.[1] With regard to the amino acid sequence, in difference from Pup and Ubiquitin, UBact does not contain a di-glycine motif at its C-terminus. Rather, it usually ends with the sequence R[T/S]G[E/Q] (see figure 3).
It took almost ten years since the discovery of Pup in 2008,[8] to identify UBact. This is probably due to the difference between Pup and UBact amino acids sequences, and because very few bacteria from the five phyla where UBact is found have been sequenced.[1]
Bacteria from the phyla where UBact is found interact with humans,[9][10] and are found in the human gut microbiota.[11] In marine systems, the most frequently encountered nitrogen-oxidizing bacteria are related to the UBact encoding Nitrospina gracilis [12] From the knowledge accumulated about the Pup-proteasome system and its importance in bacterial durability and disease causing ability,[13][14] the homologous UBact-proteasome system is expected to have similar impact on the gram-negative bacteria where it is found. In addition to humans, animals such livestock and fish that eat from the ground or swim in water are expected to be constantly exposed to UBact containing bacteria in the soil and water respectively.
From evolutionary perspective, the finding of the UBact-proteasome system in gram-negtive bacteria suggests that either the Pup/UBact-proteasome systems evolved in bacteria prior to the split into gram positive and negative clades over 3000 million years ago[15] or, that these systems were acquired by different bacterial lineages through horizontal gene transfer(s) from a third, yet unknown, organism. In support of the second possibility, two UBact loci were found in the genome of an uncultured anaerobic methanotrophic Archaeon (ANME-1;locus CBH38808.1 and locus CBH39258.1). More possibilities exist.
Update: UBact is also found in the gram-negative bacterial phylum Gemmatimonadetes (e.g., A0A2E8WA32, A0A2E3J6F7, A0A2E7JSE3) and in the candidate phylum Latescibacteria (previously known as WS3; e.g., A0A3D2RHP4, A0A3D5FTR6, A0A3D4H075, and A0A3B8MMW3).
References
- Lehmann, G.; Udasin, R.G.; Livneh, I.; Ciechanover, A. (2017). "Identification of UBact, a ubiquitin-like protein, along with other homologous components of a conjugation system and the proteasome in different gram-negative bacteria". Biochem Biophys Res Commun. 483 (3): 946–950. doi:10.1016/j.bbrc.2017.01.037. PMID 28087277.
- Delley, C.L.; Mueller, A.U.; Ziemski, M.; Weber-Ban, E. (2017). "Prokaryotic Ubiquitin-Like Protein and Its Ligase/Deligase Enzymes". J Mol Biol. 429 (22): 3486–3499. doi:10.1016/j.jmb.2017.04.020. hdl:20.500.11850/191988. PMID 28478282.
- Vijay-Kumar, S.; Bugg, C.E.; Cook, W.J. (1987). "Structure of ubiquitin refined at 1.8 A resolution". J Mol Biol. 194 (3): 531–544. doi:10.1016/0022-2836(87)90679-6. PMID 3041007.
- Chen, X.; Solomon, W.C.; Kang, Y.; Cerda-Maira, F.; Darwin, K.H.; Walters, K.J. (2009). "Prokaryotic ubiquitin-like protein pup is intrinsically disordered". J Mol Biol. 392 (1): 208–217. doi:10.1016/j.jmb.2009.07.018. PMC 2734869. PMID 19607839.
- Liao, S.; Shang, Q.; Zhang, X.; Xu, C.; Tu, X. (2009). "Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein". Biochem. J. 422 (2): 207–215. doi:10.1042/BJ20090738. PMID 19580545.
- "Ubiquitin-like protein Pup [Leptospirillum ferriphilum] - Protein - NCBI".
- "Ubiquitin-like protein UBact [Leptospirillum ferriphilum] - Protein - NCBI".
- Pearce, M. J.; Mintseris, J.; Ferreyra, J.; Gygi, S. P.; Darwin, K. H. (2008). "Ubiquitin-Like Protein Involved in the Proteasome Pathway of Mycobacterium tuberculosis". Science. 322 (5904): 1104–1107. Bibcode:2008Sci...322.1104P. doi:10.1126/science.1163885. PMC 2698935. PMID 18832610.
- Kowarsky, Mark; Camunas-Soler, Joan; Kertesz, Michael; Vlaminck, Iwijn De; Koh, Winston; Pan, Wenying; Martin, Lance; Neff, Norma F.; Okamoto, Jennifer (2017-09-05). "Numerous uncharacterized and highly divergent microbes which colonize humans are revealed by circulating cell-free DNA". Proceedings of the National Academy of Sciences. 114 (36): 9623–9628. doi:10.1073/pnas.1707009114. ISSN 0027-8424. PMC 5594678. PMID 28830999.
- Drancourt, M.; Prebet, T.; Aghnatios, R.; Edouard, S.; Cayrou, C.; Henry, M.; Blaise, D.; Raoult, D. (2014). "Planctomycetes DNA in febrile aplastic patients with leukemia, rash, diarrhea, and micronodular pneumonia". J Clin Microbiol. 52 (9): 3453–3455. doi:10.1128/JCM.01207-14. PMC 4313204. PMID 24920769.
- Dubourg, G.; Lagier, J.C.; Armougom, F.; Robert, C.; Audoly, G.; Papazian, L.; Raoult, D. (2013). "High-level colonisation of the human gut by Verrucomicrobia following broad-spectrum antibiotic treatment". Int J Antimicrob Agents. 41 (2): 149–155. doi:10.1016/j.ijantimicag.2012.10.012. PMID 23294932.
- Lücker, S.; Nowka, B.; Rattei, T.; Spieck, E.; Daims, H. (2013). "The Genome of Nitrospina gracilis Illuminates the Metabolism and Evolution of the Major Marine Nitrite Oxidizer". Front. Microbiol. 4 (27): eCollection. doi:10.3389/fmicb.2013.00027. PMC 3578206. PMID 23439773.
- Becker, S.H.; Darwin, K.H. (2016). "Bacterial Proteasomes: Mechanistic and Functional Insights". Microbiol Mol Biol Rev. 81 (1): e00036–16. doi:10.1128/MMBR.00036-16. PMC 5312241. PMID 27974513.
- Elharar, Y.; Roth, Z.; Hecht, N.; Rotkopf, R.; Khalaila, I.; Gur, E. (2016). "Posttranslational regulation of coordinated enzyme activities in the Pup-proteasome system". Proc Natl Acad Sci U S A. 113 (12): E1605–E1614. Bibcode:2016PNAS..113E1605E. doi:10.1073/pnas.1525185113. PMC 4812726. PMID 26951665.
- Marin, J.; Battistuzzi, F.U.; Brown, A.C.; Hedges, S.B. (2017). "The Timetree of Prokaryotes: New Insights into Their Evolution and Speciation". Mol Biol Evol. 34 (2): 437–446. doi:10.1093/molbev/msw245. PMID 27965376.