Topologically associating domain
A topologically associating domain (TAD) is a self-interacting genomic region, meaning that DNA sequences within a TAD physically interact with each other more frequently than with sequences outside the TAD.[1] The median size of a TAD in mouse cells is 880 kb, and they have similar sizes in non-mammalian species.[2] Boundaries at both side of the these domains are conserved between different mammalian cell types and even across species[2] and are highly enriched with CCCTC-binding factor (CTCF) and cohesin binding sites.[1] In addition, some types of genes (such as transfer RNA genes and housekeeping genes) appear near TAD boundaries more often than would be expected by chance.[3][4]
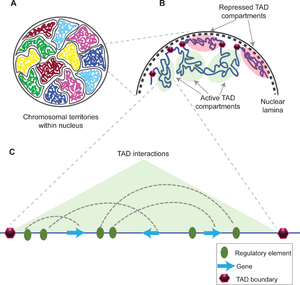
The functions of TADs are not fully understood and still is a matter of debate. Most of the studies indicate TADs regulate gene expression by limiting the enhancer-promoter interaction to each TAD,[5] however, a recent study uncouples TAD organization and gene expression.[6] Disruption of TAD boundaries are found to be associated with wide range of diseases such as cancer,[7][8][9] variety of limb malformations such as synpolydactyly, Cooks syndrome, and F-syndrome,[10] and number of brain disorders like Hypoplastic corpus callosum and Adult-onset demyelinating leukodystrophy.[10]
The mechanisms underlying TAD formation are also complex and not yet fully elucidated, though a number of protein complexes and DNA elements are associated with TAD boundaries. However, the handcuff model and the loop extrusion model are described to describe the TAD formation by the aid of CTCF and cohesin proteins.[11] Furthermore, it has been proposed that the stiffness of TAD boundaries itself could cause the domain insulation and TAD formation.[11]
Discovery and diversity
TADs are defined as regions whose DNA sequences preferentially contact each other. They were discovered in 2012 using chromosome conformation capture techniques including Hi-C.[3][12][4] They have been shown to be present in multiple species,[13] including fruit flies (Drosophila),[14] mouse,[3] plants, fungi and human[4] genomes. In bacteria, they are referred to as Chromosomal Interacting Domains (CIDs).[13]
Analytical tools and databases
TAD locations are defined by applying an algorithm to Hi-C data. For example, TADs are often called according to the so-called "directionality index".[4] The directionality index is calculated for individual 40kb bins, by collecting the reads that fall in the bin, and observing whether their paired reads map upstream or downstream of the bin (read pairs are required to span no more than 2Mb). A positive value indicates that more read pairs lie downstream than upstream, and a negative value indicates the reverse. Mathematically, the directionality index is a signed chi-square statistic.
The development of 3D genome browsers and databases such as The 3D Genome Browser,[15] 3DIV,[16] 3D-GNOME[17], and TADKB[18] have enabled us to visualize the TAD organization of regions of interest in different cell types.
Mechanisms of formation
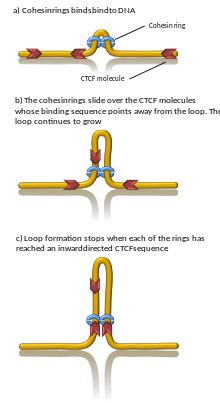
A number of proteins are known to be associated with TAD formation including the protein CTCF and the protein complex cohesin.[1] It is also unknown what components are required at TAD boundaries; however, in mammalian cells, it has been shown that these boundary regions have comparatively high levels of CTCF binding. In addition, some types of genes (such as transfer RNA genes and housekeeping genes) appear near TAD boundaries more often than would be expected by chance.[3][4]
Computer simulations have shown that chromatin loop extrusion driven by transcription generated supercoiling ensures that cohesin relocalizes quickly and loops grow with reasonable speed and in a good direction. In addition, the supercoiling-driven loop extrusion mechanism is consistent with earlier explanations proposing why TADs flanked by convergent CTCF binding sites form more stable chromatin loops than TADs flanked by divergent CTCF binding sites. In this model, the supercoiling also stimulates enhancer promoter contacts and it is proposed that transcription of eRNA sends the first wave of supercoiling that can activate mRNA transcription in a given TAD.[19][20] Computational models also showed that cohesin rings act like a very efficient molecular comb, pushing knots and entanglements such as in catenanes towards border of TADs where these are removed by the action of topoisomerases. Consistently, removal of entanglements during loop extrusion also increases degree of segregation between chromosomes.[21] However, proof for DNA loop-extrusion is so far limited to condensin (cohesin's sister protein complex) only.[22]
Properties
Conservation
TADs have been reported to be relatively constant between different cell types (in stem cells and blood cells, for example), and even between species in specific cases.[23][24]
Relationship with promoter-enhancer contacts
The majority of observed interactions between promoters and enhancers do not cross TAD boundaries. Removing a TAD boundary (for example, using CRISPR to delete the relevant region of the genome) can allow new promoter-enhancer contacts to form. This can affect gene expression nearby - such misregulation has been shown to cause limb malformations (e.g. polydactyly) in humans and mice.[23]
Computer simulations have shown that transcription-induced supercoiling of chromatin fibres can explain how TADs are formed and how they can assure very efficient interactions between enhancers and their cognate promoters located in the same TAD.[20]
Relationship with other structural features of the genome
Replication timing domains have been shown to be associated with TADs as their boundary is co localized with the boundaries of TADs that are located at either sides of compartments.[25] Insulated neighborhoods, DNA loops formed by CTCF/cohesin-bound regions, are proposed to functionally underlie TADs.[26]
Role in disease
Disruption of TAD boundaries can affect the expression of nearby genes, and this can cause disease.[27]
For example, genomic structural variants that disrupt TAD boundaries have been reported to cause developmental disorders such as human limb malformations.[28][29][30] Additionally, several studies have provided evidence that the disruption or rearrangement of TAD boundaries can provide growth advantages to certain cancers, such as T-cell acute lymphoblastic leukemia (T-ALL),[31] gliomas,[32] and lung cancer.[33]
Lamina-associated domains

Lamina-associated domains (LADs) are parts of the chromatin that heavily interact with the lamina, a network-like structure at the inner membrane of the nucleus.[34] LADs consist mostly of transcriptionally silent chromatin, being enriched with trimethylated Lys27 on histone H3, which is a common posttranslational histone modification of heterochromatin.[35] LADs have CTCF-binding sites at their periphery.[34]
See also
References
- Pombo A, Dillon N (April 2015). "Three-dimensional genome architecture: players and mechanisms". Nature Reviews. Molecular Cell Biology. 16 (4): 245–57. doi:10.1038/nrm3965. PMID 25757416.
- Yu M, Ren B (October 2017). "The Three-Dimensional Organization of Mammalian Genomes". Annual Review of Cell and Developmental Biology. 33: 265–289. doi:10.1146/annurev-cellbio-100616-060531. PMC 5837811. PMID 28783961.
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. (April 2012). "Spatial partitioning of the regulatory landscape of the X-inactivation centre". Nature. 485 (7398): 381–5. Bibcode:2012Natur.485..381N. doi:10.1038/nature11049. PMC 3555144. PMID 22495304.
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. (April 2012). "Topological domains in mammalian genomes identified by analysis of chromatin interactions". Nature. 485 (7398): 376–80. Bibcode:2012Natur.485..376D. doi:10.1038/nature11082. PMC 3356448. PMID 22495300.
- Krijger PH, de Laat W (December 2016). "Regulation of disease-associated gene expression in the 3D genome". Nature Reviews. Molecular Cell Biology. 17 (12): 771–782. doi:10.1038/nrm.2016.138. PMID 27826147.
- Ghavi-Helm Y, Jankowski A, Meiers S, Viales RR, Korbel JO, Furlong EE (August 2019). "Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression". Nature Genetics. 51 (8): 1272–1282. doi:10.1038/s41588-019-0462-3. PMID 31308546.
- Corces MR, Corces VG (February 2016). "The three-dimensional cancer genome". Current Opinion in Genetics & Development. 36: 1–7. doi:10.1016/j.gde.2016.01.002. PMC 4880523. PMID 26855137.
- Valton AL, Dekker J (February 2016). "TAD disruption as oncogenic driver". Current Opinion in Genetics & Development. 36: 34–40. doi:10.1016/j.gde.2016.03.008. PMC 4880504. PMID 27111891.
- Achinger-Kawecka J, Clark SJ (January 2017). "Disruption of the 3D cancer genome blueprint". Epigenomics. 9 (1): 47–55. doi:10.2217/epi-2016-0111. PMID 27936932.
- Spielmann M, Lupiáñez DG, Mundlos S (July 2018). "Structural variation in the 3D genome". Nature Reviews. Genetics. 19 (7): 453–467. doi:10.1038/s41576-018-0007-0. hdl:21.11116/0000-0003-610A-5. PMID 29692413.
- Dixon JR, Gorkin DU, Ren B (June 2016). "Chromatin Domains: The Unit of Chromosome Organization". Molecular Cell. 62 (5): 668–80. doi:10.1016/j.molcel.2016.05.018. PMC 5371509. PMID 27259200.
- de Laat W, Duboule D (October 2013). "Topology of mammalian developmental enhancers and their regulatory landscapes". Nature. 502 (7472): 499–506. Bibcode:2013Natur.502..499D. doi:10.1038/nature12753. PMID 24153303.
- Szabo Q, Bantignies F, Cavalli G (April 2019). "Principles of genome folding into topologically associating domains". Science Advances. 5 (4): eaaw1668. doi:10.1126/sciadv.aaw1668. PMC 6457944. PMID 30989119.
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, et al. (February 2012). "Three-dimensional folding and functional organization principles of the Drosophila genome". Cell. 148 (3): 458–72. doi:10.1016/j.cell.2012.01.010. PMID 22265598.
- Wang Y, Song F, Zhang B, Zhang L, Xu J, Kuang D, et al. (October 2018). "The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions". Genome Biology. 19 (1): 151. doi:10.1186/s13059-018-1519-9. PMC 6172833. PMID 30286773.
- Yang D, Jang I, Choi J, Kim MS, Lee AJ, Kim H, et al. (January 2018). "3DIV: A 3D-genome Interaction Viewer and database". Nucleic Acids Research. 46 (D1): D52–D57. doi:10.1093/nar/gkx1017. PMC 5753379. PMID 29106613.
- Szalaj P, Michalski PJ, Wróblewski P, Tang Z, Kadlof M, Mazzocco G, et al. (July 2016). "3D-GNOME: an integrated web service for structural modeling of the 3D genome". Nucleic Acids Research. 44 (W1): W288-93. doi:10.1093/nar/gkw437. PMC 4987952. PMID 27185892.
- Liu, T., Porter, J., Zhao, C. et al. TADKB: Family classification and a knowledge base of topologically associating domains. BMC Genomics 20, 217 (2019). https://doi.org/10.1186/s12864-019-5551-2
- Racko D, Benedetti F, Dorier J, Stasiak A (February 2018). "Transcription-induced supercoiling as the driving force of chromatin loop extrusion during formation of TADs in interphase chromosomes". Nucleic Acids Research. 46 (4): 1648–1660. doi:10.1093/nar/gkx1123. PMC 5829651. PMID 29140466.

- Racko D, Benedetti F, Dorier J, Stasiak A (January 2019). "Are TADs supercoiled?". Nucleic Acids Research. 47 (2): 521–532. doi:10.1093/nar/gky1091. PMC 6344874. PMID 30395328.
- Racko D, Benedetti F, Goundaroulis D, Stasiak A (October 2018). "Chromatin Loop Extrusion and Chromatin Unknotting". Polymers. 10 (10): 1126–1137. doi:10.3390/polym10101126. PMC 6403842. PMID 30961051.
- Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, Dekker C (April 2018). "Real-time imaging of DNA loop extrusion by condensin". Science. 360 (6384): 102–105. doi:10.1126/science.aar7831. PMC 6329450. PMID 29472443.
- Jost D, Vaillant C, Meister P (February 2017). "Coupling 1D modifications and 3D nuclear organization: data, models and function". Current Opinion in Cell Biology. 44: 20–27. doi:10.1016/j.ceb.2016.12.001. PMID 28040646.
- Yang Y, Zhang Y, Ren B, Dixon JR, Ma J (June 2019). "Comparing 3D Genome Organization in Multiple Species Using Phylo-HMRF". Cell Systems. 8 (6): 494–505.e14. doi:10.1016/j.cels.2019.05.011. PMC 6706282. PMID 31229558.
- Marchal C, Sima J, Gilbert DM (December 2019). "Control of DNA replication timing in the 3D genome". Nature Reviews. Molecular Cell Biology. 20 (12): 721–737. doi:10.1038/s41580-019-0162-y. PMID 31477886.
- Ji X, Dadon DB, Powell BE, Fan ZP, Borges-Rivera D, Shachar S, et al. (February 2016). "3D Chromosome Regulatory Landscape of Human Pluripotent Cells". Cell Stem Cell. 18 (2): 262–75. doi:10.1016/j.stem.2015.11.007. PMC 4848748. PMID 26686465.
- Lupiáñez DG, Spielmann M, Mundlos S (April 2016). "Breaking TADs: How Alterations of Chromatin Domains Result in Disease". Trends in Genetics. 32 (4): 225–237. doi:10.1016/j.tig.2016.01.003. hdl:11858/00-001M-0000-002E-1D1D-D. PMID 26862051.
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, et al. (May 2015). "Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions". Cell. 161 (5): 1012–1025. doi:10.1016/j.cell.2015.04.004. PMC 4791538. PMID 25959774.
- Angier N (2017-01-09). "A Family's Shared Defect Sheds Light on the Human Genome". The New York Times.
- Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, et al. (October 2016). "Formation of new chromatin domains determines pathogenicity of genomic duplications". Nature. 538 (7624): 265–269. Bibcode:2016Natur.538..265F. doi:10.1038/nature19800. PMID 27706140.
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, et al. (March 2016). "Activation of proto-oncogenes by disruption of chromosome neighborhoods". Science. 351 (6280): 1454–1458. Bibcode:2016Sci...351.1454H. doi:10.1126/science.aad9024. PMC 4884612. PMID 26940867.
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al. (January 2016). "Insulator dysfunction and oncogene activation in IDH mutant gliomas". Nature. 529 (7584): 110–4. Bibcode:2016Natur.529..110F. doi:10.1038/nature16490. PMC 4831574. PMID 26700815.
- Weischenfeldt J, Dubash T, Drainas AP, Mardin BR, Chen Y, Stütz AM, et al. (January 2017). "Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking". Nature Genetics. 49 (1): 65–74. doi:10.1038/ng.3722. PMC 5791882. PMID 27869826.
- Gonzalez-Sandoval A, Gasser SM (August 2016). "On TADs and LADs: Spatial Control Over Gene Expression". Trends in Genetics. 32 (8): 485–495. doi:10.1016/j.tig.2016.05.004. PMID 27312344.
- Li M, Liu GH, Izpisua Belmonte JC (July 2012). "Navigating the epigenetic landscape of pluripotent stem cells". Nature Reviews. Molecular Cell Biology. 13 (8): 524–35. doi:10.1038/nrm3393. PMID 22820889.