TMEM106A
TMEM106A is a gene that encodes the transmembrane protein 106A (TMEM106A) in Homo sapiens.[5] It is located at 17q21.31 on the plus strand next to cancer-related genes NBR1 and BRCA1.[5][6] The TMEM106A gene contains a domain of unknown function, DUF1356.[5]
Protein structure
The TMEM106A protein has a molecular weight of 28.9 kdal. It has 262 amino acids, 240 of which are in the domain of function.[5] The protein has a transmembrane region.[7] There is evidence for a secondary transmembrane region in humans but this region is not conserved in related orthologs.[8] The protein does not contain a peptide signal protein.[9] The protein structure contains a similar proportion of alpha-helix and beta-strand secondary structures (this does not include transmembrane structures).[10][11]
There are several areas for post-translational modification for TMEM106A including:
Homology
Paralogs
The TMEM106A gene has two paralogs: TMEM106B and TMEM106C. These paralogs belong to the gene family pfam07092, which belongs to the DUF1356 superfamily. This family consists of several mammalian proteins that are around 250 amino acids in length.[15] TMEM106B and TMEM106C are conserved in invertebrates to mammals.
| Protein | Accession Number | Amino Acids | Identity Percent | Highest Expression |
|---|---|---|---|---|
| TMEM106A | AAI46977 | 262 | 100 | Kidney [16] |
| TMEM106B | NP_001127704 | 274 | 43 | Ubiquitous [17] |
| TMEM106C | AAI07793 | 231 | 36 | Ubiquitous [18] |
Orthologs
The TMEM106A gene has been found in only the Chordate phylum.[20] Of the three subphyla, TMEM106A is most commonly found in Vertebrata and has also been found in select members of Tunicata, which are invertebrate marine filter feeders. This phylum split occurred 722.5 million years ago.[21] TMEM106A has not been seen in bacteria, plants, or fungi.
| Organism | Common Name | Accession Number | Amino Acids | Identity Percent | Notes |
|---|---|---|---|---|---|
| Homo sapiens | Human | AAI46977.1 | 262 | 100 | Mammal |
| Pan troglodytes | Chimpanzee | XP_001154896.2 | 262 | 99.2 | Mammal |
| Pongo abelii | Orangutan | XP_002827523.1 | 262 | 96.2 | Mammal |
| Callithrix jacchus | Marmoset | XP_002748067.1 | 262 | 90.5 | Mammal |
| Canis lupus familiaris | Dog | XP_548074.2 | 262 | 84.8 | Mammal |
| Mus musculus | Mouse | AAH22145.1 | 261 | 66.4 | Mammal |
| Xenopus borealis | Marsabit Clawed Frog | ACC55056.1 | 262 | 59.5 | Reptile |
| Danio rerio | Zebrafish | AAH50177.1 | 282 | 34.5 | Fish |
| Oikopleura dioica | Sea-squirt | CBY08060.1 | 249 | 27.8 | Invertebrate |
Expression
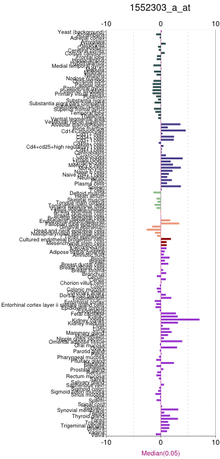
TMEM106A is expressed in several human tissues. The tissues with highest expression are uterus, kidneys, small intestine, and stomach.[16][22] EST profiles for orthologs show expression is conserved with greatest expression in kidneys and lesser expression in several other areas.[23] Some tissues never show expression including: muscle, adipose tissue, and bone.
Gene neighborhood
In Homo sapiens, TMEM106A is located next to NBR1, a gene identified as an ovarian tumor antigen monitored in ovarian cancer.[24] It is also located near BRCA1, a breast cancer tumor suppressor gene.[25] The first 140 amino acids of the TMEM106A protein, including portions of DUF1356 and a transmembrane region, are deleted along with BRCA1 during early-onset breast cancer.[26]

References
- GRCh38: Ensembl release 89: ENSG00000184988 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000034947 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: TMEM106A transmembrane protein 106A".
- "Genecards: TMEM106A transmembrane protein 106A".
- Nakai K, Horton P (January 1999). "PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization". Trends Biochem. Sci. 24 (1): 34–6. doi:10.1016/S0968-0004(98)01336-X. PMID 10087920.
- Persson B, Argos P (March 1994). "Prediction of transmembrane segments in proteins utilising multiple sequence alignments". J. Mol. Biol. 237 (2): 182–92. doi:10.1006/jmbi.1994.1220. PMID 8126732.
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (January 1997). "Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites". Protein Eng. 10 (1): 1–6. doi:10.1093/protein/10.1.1. PMID 9051728.
- Garnier J, Osguthorpe DJ, Robson B (March 1978). "Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins". J. Mol. Biol. 120 (1): 97–120. doi:10.1016/0022-2836(78)90297-8. PMID 642007.
- Chou PY, Fasman GD (1978). Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. Advances in Enzymology - and Related Areas of Molecular Biology. 47. pp. 45–148. doi:10.1002/9780470122921.ch2. ISBN 9780470122921. PMID 364941.
- Blom N, Gammeltoft S, Brunak S (December 1999). "Sequence and structure-based prediction of eukaryotic protein phosphorylation sites". J. Mol. Biol. 294 (5): 1351–62. doi:10.1006/jmbi.1999.3310. PMID 10600390.
- Gupta R, Jung E, Brunak S (2004). "Prediction of N-glycosylation sites in human proteins". Cite journal requires
|journal=(help) - Johansen MB, Kiemer L, Brunak S (September 2006). "Analysis and prediction of mammalian protein glycation". Glycobiology. 16 (9): 844–53. CiteSeerX 10.1.1.128.831. doi:10.1093/glycob/cwl009. PMID 16762979.
- "NCBI Conserved Domains: DUF1356".
- "EST profile: TMEM106A transmembrane protein 106A"..
- "EST profile: TMEM106B transmembrane protein 106B"..
- "EST profile: TMEM106C transmembrane protein 106C"..
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, Su AI (2009). "BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources". Genome Biol. 10 (11): R130. doi:10.1186/gb-2009-10-11-r130. PMC 3091323. PMID 19919682.
- "NCBI Homologene: TMEM106A".
- Hedges SB, Dudley J, Kumar S (December 2006). "TimeTree: a public knowledge-base of divergence times among organisms". Bioinformatics. 22 (23): 2971–2. doi:10.1093/bioinformatics/btl505. PMID 17021158.
- "GEO Profiles: TMEM106A transmembrane protein 106A".
- "EST profiles"..
- Whitehouse C, Chambers J, Howe K, Cobourne M, Sharpe P, Solomon E (January 2002). "NBR1 interacts with fasciculation and elongation protein zeta-1 (FEZ1) and calcium and integrin binding protein (CIB) and shows developmentally restricted expression in the neural tube". Eur. J. Biochem. 269 (2): 538–45. doi:10.1046/j.0014-2956.2001.02681.x. PMID 11856312.
- Garcia-Casado Z, Romero I, Fernandez-Serra A, Rubio L, Llopis F, Garcia A, Llombart P, Lopez-Guerrero JA (2011). "A de novo complete BRCA1 gene deletion identified in a Spanish woman with early bilateral breast cancer". BMC Med. Genet. 12: 134. doi:10.1186/1471-2350-12-134. PMC 3207938. PMID 21989022.
- del Valle J, Feliubadaló L, Nadal M, Teulé A, Miró R, Cuesta R, Tornero E, Menéndez M, Darder E, Brunet J, Capellà G, Blanco I, Lázaro C (August 2010). "Identification and comprehensive characterization of large genomic rearrangements in the BRCA1 and BRCA2 genes". Breast Cancer Res. Treat. 122 (3): 733–43. doi:10.1007/s10549-009-0613-9. PMID 19894111.
External links
- Feric M, Zhao B, Hoffert JD, Pisitkun T, Knepper MA (April 2011). "Large-scale phosphoproteomic analysis of membrane proteins in renal proximal and distal tubule". Am. J. Physiol., Cell Physiol. 300 (4): C755–70. doi:10.1152/ajpcell.00360.2010. PMC 3074622. PMID 21209370.