TARDBP
TAR DNA-binding protein 43 (TDP-43, transactive response DNA binding protein 43 kDa), is a protein that in humans is encoded by the TARDBP gene.[5]
Structure
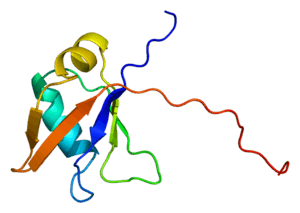
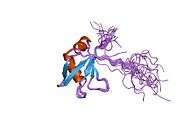
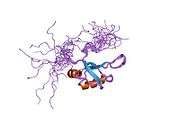
TDP-43 is 414 amino acid residues long. It consists of 4 domains: an N-terminal domain spanning residues 1-76 (NTD) with a well-defined fold that has been shown to form a dimer or oligomer;[6][7] 2 highly conserved folded RNA recognition motifs spanning residues 106-176 (RRM1) and 191-259 (RRM2), respectively, required to bind target RNA and DNA;[8] an unstructured C-terminal domain encompassing residues 274-414 (CTD), which contains a glycine-rich region, is involved in protein-protein interactions, and harbors most of the mutations associated with familial amyotrophic lateral sclerosis.[9]
The entire protein devoid of large solubilising tags has been recently purified.[10] The full-length protein is a dimer.[10] The dimer is formed due to a self-interaction between two NTD domains,[6][7] where the dimerisation can be propagated to form higher-order oligomers.[6]
The protein sequence also has a nuclear localization signal (NLS, residues 82–98), a nuclear export signal (NES residues 239–250) and 3 putative caspase-3 cleavage sites (residues 13, 89, 219).[10]
Function
TDP-43 is a transcriptional repressor that binds to chromosomally integrated TAR DNA and represses HIV-1 transcription. In addition, this protein regulates alternate splicing of the CFTR gene. In particular, TDP-43 is a splicing factor binding to the intron8/exon9 junction of the CFTR gene and to the intron2/exon3 region of the apoA-II gene.[11] A similar pseudogene is present on chromosome 20.[12]
TDP-43 has been shown to bind both DNA and RNA and have multiple functions in transcriptional repression, pre-mRNA splicing and translational regulation. Recent work has characterized the transcriptome-wide binding sites revealing that thousands of RNAs are bound by TDP-43 in neurons.[13]
TDP-43 was originally identified as a transcriptional repressor that binds to chromosomally integrated trans-activation response element (TAR) DNA and represses HIV-1 transcription.[5] It was also reported to regulate alternate splicing of the CFTR gene and the apoA-II gene.[14][15]
In spinal motor neurons TDP-43 has also been shown in humans to be a low molecular weight neurofilament (hNFL) mRNA-binding protein.[16] It has also shown to be a neuronal activity response factor in the dendrites of hippocampal neurons suggesting possible roles in regulating mRNA stability, transport and local translation in neurons.[17]
Recently, it has been demonstrated that zinc ions are able to induce aggregation of endogenous TDP-43 in cells.[18] Moreover, zinc could bind to RNA binding domain of TDP-43 and induce the formation of amyloid-like aggregates in vitro.[19]
DNA repair
TDP-43 protein is a key element of the non-homologous end joining (NHEJ) enzymatic pathway that repairs DNA double-strand breaks (DSBs) in pluripotent stem cell-derived motor neurons.[20] TDP-43 is rapidly recruited to DSBs where it acts as a scaffold for the further recruitment of the XRCC4-DNA ligase protein complex that then acts to seal the DNA breaks. In TDP-43 depleted human neural stem cell-derived motor neurons, as well as in sporadic ALS patients’ spinal cord specimens there is significant DSB accumulation and reduced levels of NHEJ.[20]
Clinical significance
A hyper-phosphorylated, ubiquitinated and cleaved form of TDP-43—known as pathologic TDP43—is the major disease protein in ubiquitin-positive, tau-, and alpha-synuclein-negative frontotemporal dementia (FTLD-TDP, previously referred to as FTLD-U[21]) and in amyotrophic lateral sclerosis (ALS).[22][23] Elevated levels of the TDP-43 protein have also been identified in individuals diagnosed with chronic traumatic encephalopathy, and has also been associated with ALS leading to the inference that athletes who have experienced multiple concussions and other types of head injury are at an increased risk for both encephalopathy and motor neuron disease (ALS).[24] Abnormalities of TDP-43 also occur in an important subset of Alzheimer's disease patients, correlating with clinical and neuropathologic features indexes.[25] Misfolded TDP-43 is found in the brains of older adults over age 85 with limbic-predominant age-related TDP-43 encephalopathy, (LATE), a form of dementia.
HIV-1, the causative agent of acquired immunodeficiency syndrome (AIDS), contains an RNA genome that produces a chromosomally integrated DNA during the replicative cycle. Activation of HIV-1 gene expression by the transactivator "Tat" is dependent on an RNA regulatory element (TAR) located "downstream" (i.e. to-be transcribed at a later point in time) of the transcription initiation site.
Mutations in the TARDBP gene are associated with neurodegenerative disorders including frontotemporal lobar degeneration and amyotrophic lateral sclerosis (ALS).[26] In particular, the TDP-43 mutants M337V and Q331K are being studied for their roles in ALS.[27][28][29] Cytoplasmic TDP-43 pathology is the dominant histopathological feature of multisystem proteinopathy.[30] The N-terminal domain, which contributes importantly to the aggregation of the C-terminal region, has a novel structure with two negatively charged loops.[31] A recent study has demonstrated that cellular stress can trigger the abnormal cytoplasmic mislocalisation of TDP-43 in spinal motor neurons in vivo, providing insight into how TDP-43 pathology may develop in sporadic ALS patients.[32]
References
- GRCh38: Ensembl release 89: ENSG00000120948 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000041459 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Ou SH, Wu F, Harrich D, García-Martínez LF, Gaynor RB (June 1995). "Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs". Journal of Virology. 69 (6): 3584–96. PMC 189073. PMID 7745706.
- Afroz T, Hock EM, Ernst P, Foglieni C, Jambeau M, Gilhespy L, Laferriere F, Maniecka Z, Plückthun A, Mittl P, Paganetti P, Allain FH, Polymenidou M (June 2017). "Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation". Nature Communications. 8: 45. doi:10.1038/s41467-017-00062-0. PMC 5491494. PMID 28663553.
- Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, Reeb AN, Nourse A, Ramirez Montero D, Ryan VH, Rohatgi R, Shewmaker F, Naik MT, Mittag T, Ayala YM, Fawzi NL (March 1, 2018). "A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing". EMBO Journal. 37: e97452. doi:10.15252/embj.201797452. PMC 5830921. PMID 29438978.
- Lukavsky PJ, Daujotyte D, Tollervey JR, Ule J, Stuani C, Buratti E, Baralle FE, Damberger FF, Allain FH (December 2013). "Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43". Nature Struct Mol Biol. 20: 1443. doi:10.1038/nsmb.2698. PMID 2424061.
- Conicella AE, Zerze GH, Mittal J, Fawzi NL (6 September 2016). "ALS Mutations Disrupt Phase Separation Mediated by α-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain". Structure. 24: 1537–49. doi:10.1016/j.str.2016.07.007. PMC 5014597. PMID 27545621.
- Vivoli Vega M, Nigro A, Luti S, Capitini C, Fani G, Gonnelli L, Boscaro F, Chiti F (October 2019). "Isolation and characterization of soluble human full-length TDP-43 associated with neurodegeneration". FASEB J. 33: 10780–93. doi:10.1096/fj.201900474R. PMID 31287959.
- Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS (April 2009). "Structural insights into TDP-43 in nucleic-acid binding and domain interactions". Nucleic Acids Research. 37 (6): 1799–808. doi:10.1093/nar/gkp013. PMC 2665213. PMID 19174564.
- Gene Result
- Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, Moore MJ, Yu G (January 2011). "Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes". The Journal of Biological Chemistry. 286 (2): 1204–15. doi:10.1074/jbc.M110.190884. PMC 3020728. PMID 21051541.
- Buratti E, Baralle FE (September 2001). "Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9". The Journal of Biological Chemistry. 276 (39): 36337–43. doi:10.1074/jbc.M104236200. PMID 11470789.
- Mercado PA, Ayala YM, Romano M, Buratti E, Baralle FE (2005-10-12). "Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene". Nucleic Acids Research. 33 (18): 6000–10. doi:10.1093/nar/gki897. PMC 1270946. PMID 16254078.
- Strong MJ, Volkening K, Hammond R, Yang W, Strong W, Leystra-Lantz C, Shoesmith C (June 2007). "TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein". Molecular and Cellular Neurosciences. 35 (2): 320–7. doi:10.1016/j.mcn.2007.03.007. PMID 17481916.
- Wang IF, Wu LS, Chang HY, Shen CK (May 2008). "TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor". Journal of Neurochemistry. 105 (3): 797–806. doi:10.1111/j.1471-4159.2007.05190.x. PMID 18088371.
- Caragounis A, Price KA, Soon CP, Filiz G, Masters CL, Li QX, Crouch PJ, White AR (May 2010). "Zinc induces depletion and aggregation of endogenous TDP-43". Free Radical Biology & Medicine. 48 (9): 1152–61. doi:10.1016/j.freeradbiomed.2010.01.035. PMID 20138212.
- Garnier C, Devred F, Byrne D, Puppo R, Roman AY, Malesinski S, Golovin AV, Lebrun R, Ninkina NN, Tsvetkov PO (July 2017). "Zinc binding to RNA recognition motif of TDP-43 induces the formation of amyloid-like aggregates". Scientific Reports. 7 (1): 6812. Bibcode:2017NatSR...7.6812G. doi:10.1038/s41598-017-07215-7. PMC 5533730. PMID 28754988.
- Mitra J, Guerrero EN, Hegde PM, Liachko NF, Wang H, Vasquez V, Gao J, Pandey A, Taylor JP, Kraemer BC, Wu P, Boldogh I, Garruto RM, Mitra S, Rao KS, Hegde ML (2019). "Motor neuron disease-associated loss of nuclear TDP-43 is linked to DNA double-strand break repair defects". Proc Natl Acad Sci U S A. 116: 4696–4705. doi:10.1073/pnas.1818415116. PMC 6410842. PMID 30770445.
- Mackenzie IR, Neumann M, Baborie A, Sampathu DM, Du Plessis D, Jaros E, Perry RH, Trojanowski JQ, Mann DM, Lee VM (July 2011). "A harmonized classification system for FTLD-TDP pathology". Acta Neuropathologica. 122 (1): 111–3. doi:10.1007/s00401-011-0845-8. PMC 3285143. PMID 21644037.
- Bräuer S, Zimyanin V, Hermann A (April 2018). "Prion-like properties of disease-relevant proteins in amyotrophic lateral sclerosis". Journal of Neural Transmission. 125 (4): 591–613. doi:10.1007/s00702-018-1851-y. PMID 29417336.
- Lau DH, Hartopp N, Welsh NJ, Mueller S, Glennon EB, Mórotz GM, Annibali A, Gomez-Suaga P, Stoica R, Paillusson S, Miller CC (February 2018). "Disruption of ER-mitochondria signalling in fronto-temporal dementia and related amyotrophic lateral sclerosis". Cell Death & Disease. 9 (3): 327. doi:10.1038/s41419-017-0022-7. PMC 5832427. PMID 29491392.
- Schwarz, Alan. "Study Says Brain Trauma Can Mimic A.L.S.", The New York Times, August 18, 2010. Accessed August 18, 2010.
- Tremblay C, St-Amour I, Schneider J, Bennett DA, Calon F (September 2011). "Accumulation of transactive response DNA binding protein 43 in mild cognitive impairment and Alzheimer disease". Journal of Neuropathology and Experimental Neurology. 70 (9): 788–98. doi:10.1097/nen.0b013e31822c62cf. PMC 3197017. PMID 21865887.
- Kwong LK, Neumann M, Sampathu DM, Lee VM, Trojanowski JQ (July 2007). "TDP-43 proteinopathy: the neuropathology underlying major forms of sporadic and familial frontotemporal lobar degeneration and motor neuron disease". Acta Neuropathologica. 114 (1): 63–70. doi:10.1007/s00401-007-0226-5. PMID 17492294.
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE (March 2008). "TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis". Science. 319 (5870): 1668–72. Bibcode:2008Sci...319.1668S. doi:10.1126/science.1154584. PMID 18309045.
- Gendron TF, Rademakers R, Petrucelli L (2013). "TARDBP mutation analysis in TDP-43 proteinopathies and deciphering the toxicity of mutant TDP-43". Journal of Alzheimer's Disease. 33 Suppl 1 (suppl 1): S35–45. doi:10.3233/JAD-2012-129036. PMC 3532959. PMID 22751173.
- Babić Leko, M; Župunski, V; Kirincich, J; Smilović, D; Hortobágyi, T; Hof, PR; Šimić, G (2019). "Molecular Mechanisms of Neurodegeneration Related to C9orf72 Hexanucleotide Repeat Expansion". Behavioural Neurology. 2019: 2909168. doi:10.1155/2019/2909168. PMC 6350563. PMID 30774737.
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi AS, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, Taylor JP (March 2013). "Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS". Nature. 495 (7442): 467–73. Bibcode:2013Natur.495..467K. doi:10.1038/nature11922. PMC 3756911. PMID 23455423.
- .Mompeán M, Romano V, Pantoja-Uceda D, Stuani C, Baralle FE, Buratti E, Laurents DV (April 2016). "The TDP-43 N-terminal domain structure at high resolution". The FEBS Journal. 283 (7): 1242–60. doi:10.1111/febs.13651. hdl:10261/162654. PMID 26756435.
- Svahn AJ, Don EK, Badrock AP, Cole NJ, Graeber MB, Yerbury JJ, Chung R, Morsch M (September 2018). "Nucleo-cytoplasmic transport of TDP-43 studied in real time: impaired microglia function leads to axonal spreading of TDP-43 in degenerating motor neurons". Acta Neuropathologica. 136 (3): 445–459. doi:10.1007/s00401-018-1875-2. PMC 6096729. PMID 29943193.
Further reading
- Kwong LK, Neumann M, Sampathu DM, Lee VM, Trojanowski JQ (July 2007). "TDP-43 proteinopathy: the neuropathology underlying major forms of sporadic and familial frontotemporal lobar degeneration and motor neuron disease". Acta Neuropathologica. 114 (1): 63–70. doi:10.1007/s00401-007-0226-5. PMID 17492294.
- Maruyama K, Sugano S (January 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Tokai N, Fujimoto-Nishiyama A, Toyoshima Y, Yonemura S, Tsukita S, Inoue J, Yamamota T (February 1996). "Kid, a novel kinesin-like DNA binding protein, is localized to chromosomes and the mitotic spindle". The EMBO Journal. 15 (3): 457–67. doi:10.1002/j.1460-2075.1996.tb00378.x. PMC 449964. PMID 8599929.
- Bonaldo MF, Lennon G, Soares MB (September 1996). "Normalization and subtraction: two approaches to facilitate gene discovery". Genome Research. 6 (9): 791–806. doi:10.1101/gr.6.9.791. PMID 8889548.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (October 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Hartley JL, Temple GF, Brasch MA (November 2000). "DNA cloning using in vitro site-specific recombination". Genome Research. 10 (11): 1788–95. doi:10.1101/gr.143000. PMC 310948. PMID 11076863.
- Wiemann S, Weil B, Wellenreuther R, Gassenhuber J, Glassl S, Ansorge W, Böcher M, Blöcker H, Bauersachs S, Blum H, Lauber J, Düsterhöft A, Beyer A, Köhrer K, Strack N, Mewes HW, Ottenwälder B, Obermaier B, Tampe J, Heubner D, Wambutt R, Korn B, Klein M, Poustka A (March 2001). "Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs". Genome Research. 11 (3): 422–35. doi:10.1101/gr.GR1547R. PMC 311072. PMID 11230166.
- Buratti E, Dörk T, Zuccato E, Pagani F, Romano M, Baralle FE (April 2001). "Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping". The EMBO Journal. 20 (7): 1774–84. doi:10.1093/emboj/20.7.1774. PMC 145463. PMID 11285240.
- Buratti E, Baralle FE (September 2001). "Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9". The Journal of Biological Chemistry. 276 (39): 36337–43. doi:10.1074/jbc.M104236200. PMID 11470789.
- Wang IF, Reddy NM, Shen CK (October 2002). "Higher order arrangement of the eukaryotic nuclear bodies". Proceedings of the National Academy of Sciences of the United States of America. 99 (21): 13583–8. Bibcode:2002PNAS...9913583W. doi:10.1073/pnas.212483099. PMC 129717. PMID 12361981.
- Lehner B, Sanderson CM (July 2004). "A protein interaction framework for human mRNA degradation". Genome Research. 14 (7): 1315–23. doi:10.1101/gr.2122004. PMC 442147. PMID 15231747.
- Wiemann S, Arlt D, Huber W, Wellenreuther R, Schleeger S, Mehrle A, Bechtel S, Sauermann M, Korf U, Pepperkok R, Sültmann H, Poustka A (October 2004). "From ORFeome to biology: a functional genomics pipeline". Genome Research. 14 (10B): 2136–44. doi:10.1101/gr.2576704. PMC 528930. PMID 15489336.
- Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE (November 2005). "TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing". The Journal of Biological Chemistry. 280 (45): 37572–84. doi:10.1074/jbc.M505557200. PMID 16157593.
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE (September 2005). "A human protein-protein interaction network: a resource for annotating the proteome". Cell. 122 (6): 957–68. doi:10.1016/j.cell.2005.08.029. hdl:11858/00-001M-0000-0010-8592-0. PMID 16169070.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (October 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514.
- Mehrle A, Rosenfelder H, Schupp I, del Val C, Arlt D, Hahne F, Bechtel S, Simpson J, Hofmann O, Hide W, Glatting KH, Huber W, Pepperkok R, Poustka A, Wiemann S (January 2006). "The LIFEdb database in 2006". Nucleic Acids Research. 34 (Database issue): D415–8. doi:10.1093/nar/gkj139. PMC 1347501. PMID 16381901.
External links
| Wikimedia Commons has media related to TAR DNA-binding protein 43, TDP-43. |
- GeneReviews/NCBI/NIH/UW entry on TARDBP-Related Amyotrophic Lateral Sclerosis
- Overview of all the structural information available in the PDB for UniProt: Q13148 (TAR DNA-binding protein 43) at the PDBe-KB.