Hyperbolastic functions
The hyperbolastic functions, also known as hyperbolastic growth models, are mathematical functions that are used in medical statistical modeling. These models were originally developed to capture the growth dynamics of multicellular tumor spheres, and were introduced in 2005 by Mohammad Tabatabai, David Williams, and Zoran Bursac.[1] The precision of hyperbolastic functions in modeling real world problems is somewhat due to their flexibility in their point of inflection.[1] These functions can be used in a wide variety of modeling problems such as tumor growth, stem cell proliferation, pharma kinetics, cancer growth, sigmoid activation function in neural networks, and epidemiological disease progression or regression.[1][2]
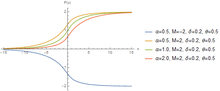

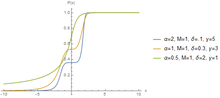



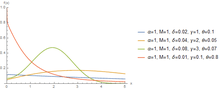
The hyperbolastic functions can model both growth and decay curves until it reaches carrying capacity. Due to their flexibility, these models have diverse applications in the medical field, with the ability to capture disease progression with an intervening treatment. As the figures indicate, the hyperbolastic functions can fit a sigmoidal curve indicating that the slowest rate occurs at the early and late stages. In addition to the presenting sigmoidal shapes, it can also accommodate biphasic situations where medical interventions slow or reverse disease progression; but, when the effect of the treatment vanishes, the disease will begin the second phase of its progression until it reaches its horizontal asymptote.
One of the main characteristics these functions have is that they cannot only fit sigmoidal shapes, but can also model biphasic growth patterns that other classical sigmoidal curves cannot adequately model. This distinguishing feature has advantageous applications in various fields including medicine, biology, economics, engineering, and computer aided system theory.[3][4][5][6]
Hyperbolastic regressions
Hyperbolastic regressions are statistical models that utilize standard hyperbolatic functions to model a dichotomous outcome variable. The purpose of binary regression is to predict a binary outcome (dependent) variable using a set of explanatory (independent) variables. Binary regression is routinely used in many areas including medical, public health, dental, and biomedical sciences. Binary regression analysis was used to predict endoscopic lesions in iron deficiency anemia.[29] In addition, binary regression was applied to differentiate between malignant and benign adnexal mass prior to surgery.[30]
The hyperbolastic regression of type I
Let be a binary outcome variable which can assume one of two mutually exclusive values, success or failure. If we code success as and failure as , the hyperbolastic success probability of type I as a function of explanatory variables is given by:
- ,
where are model parameters. The odds of success is the ratio of probability of success to the probability of failure. For hyperbolastic regression of type I, the odds of success is denoted by and expressed by the equation:
- .
The logarithm of is called the logit of Hyperbolastic of type I. The logit transformation is denoted by and can be written as:
- .
The hyperbolastic regression of type II
For the binary outcome variable , the hyperbolastic success probability of type II as a function of explanatory variables is:
- ,
For the hyperbolastic regression of type II, the odds of success is denoted by and is given by:
The logit transformation is denoted by and is given by:
References
- Tabatabai, Mohammad; Williams, David; Bursac, Zoran (2005). "Hyperbolastic growth models: Theory and application". Theoretical Biology and Medical Modelling. 2: 14. doi:10.1186/1742-4682-2-14. PMC 1084364. PMID 15799781.
- Q. Ashton Acton P. Cells-Advances in Research and Application. [Internet]. Atlanta: ScholarlyMedia LLC; 2012 [cited 2020 Apr 27]. Available from: https://public.ebookcentral.proquest.com/choice/publicfullrecord.aspx?p=4973379
- Neysens, Patricia; Messens, Winy; Gevers, Dirk; Swings, Jean; De Vuyst, Luc (2003). "Biphasic kinetics of growth and bacteriocin production with Lactobacillus amylovorus DCE 471 occur under stress conditions". Microbiology. 149 (4): 1073–1082. doi:10.1099/mic.0.25880-0. PMID 12686649.
- Chu, Charlene; Han, Christina; Shimizu, Hiromi; Wong, Bonnie (2002). "The Effect of Fructose, Galactose, and Glucose on the Induction of β-Galactosidase in Escherichia coli" (PDF). Journal of Experimental Microbiology and Immunology. 2: 1–5.
- Tabatabai, M. A.; Eby, W. M.; Singh, K. P.; Bae, S. (2013). "T model of growth and its application in systems of tumor-immunedynamics". Mathematical Biosciences and Engineering. 10 (3): 925–938. doi:10.3934/mbe.2013.10.925. PMC 4476034. PMID 23906156.
- Computer Aided Systems Theory – EUROCAST 2019. Lecture Notes in Computer Science. 12013. 2020. doi:10.1007/978-3-030-45093-9. ISBN 978-3-030-45092-2.
- Kamar SH, Msallam BS. Comparative Study between Generalized Maximum Entropy and Bayes Methods to Estimate the Four Parameter Weibull Growth Model. Journal of Probability and Statistics. 2020 Jan 14;2020:1–7.
- Roehrs T, Bogdan P, Gharaibeh B, et al. (n.d.). "Proliferative heterogeneity in stem cell populations". Live Cell Imaging Laboratory, McGowan Institute for Regenerative Medicine. Cite journal requires
|journal=(help) - Eby, Wayne M.; Tabatabai, Mohammad A.; Bursac, Zoran (2010). "Hyperbolastic modeling of tumor growth with a combined treatment of iodoacetate and dimethylsulphoxide". BMC Cancer. 10: 509. doi:10.1186/1471-2407-10-509. PMC 2955040. PMID 20863400.
- France, James; Kebreab, Ermias, eds. (2008). Mathematical Modelling in Animal Nutrition. Wallingford: CABI. ISBN 9781845933548.
- Ahmadi, H.; Mottaghitalab, M. (2007). "Hyperbolastic Models as a New Powerful Tool to Describe Broiler Growth Kinetics". Poultry Science. 86 (11): 2461–2465. doi:10.3382/ps.2007-00086. PMID 17954598.
- Choi, Taeyoung; Chin, Seongah (2014). "Novel Real-Time Facial Wound Recovery Synthesis Using Subsurface Scattering". The Scientific World Journal. 2014: 1–8. doi:10.1155/2014/965036. PMC 4146479. PMID 25197721.
- Tabatabai, M.A.; Eby, W.M.; Singh, K.P. (2011). "Hyperbolastic modeling of wound healing". Mathematical and Computer Modelling. 53 (5–6): 755–768. doi:10.1016/j.mcm.2010.10.013.
- Ko, Ung Hyun; Choi, Jongjin; Choung, Jinseung; Moon, Sunghwan; Shin, Jennifer H. (2019). "Physicochemically Tuned Myofibroblasts for Wound Healing Strategy". Scientific Reports. 9 (1): 16070. Bibcode:2019NatSR...916070K. doi:10.1038/s41598-019-52523-9. PMC 6831678. PMID 31690789.
- Barrera, Antonio; Román-Román, Patricia; Torres-Ruiz, Francisco (2020). "Diffusion Processes for Weibull-Based Models". Computer Aided Systems Theory – EUROCAST 2019. Lecture Notes in Computer Science. 12013. pp. 204–210. doi:10.1007/978-3-030-45093-9_25. ISBN 978-3-030-45092-2.
- Barrera, Antonio; Román-Román, Patricia; Torres-Ruiz, Francisco (2018). "A hyperbolastic type-I diffusion process: Parameter estimation by means of the firefly algorithm". Biosystems. 163: 11–22. doi:10.1016/j.biosystems.2017.11.001. PMID 29129822.
- Barrera, Antonio; Román-Roán, Patricia; Torres-Ruiz, Francisco (2020). "Hyperbolastic type-III diffusion process: Obtaining from the generalized Weibull diffusion process". Mathematical Biosciences and Engineering. 17 (1): 814–833. doi:10.3934/mbe.2020043. PMID 31731379.
- Barrera, Antonio; Román-Román, Patricia; Torres-Ruiz, Francisco (2020). "Two Stochastic Differential Equations for Modeling Oscillabolastic-Type Behavior". Mathematics. 8 (2): 155. doi:10.3390/math8020155.
- Stochastic Processes with Applications. 2019. doi:10.3390/books978-3-03921-729-8. ISBN 978-3-03921-729-8.
- Tabatabai, Mohammad A.; Bursac, Zoran; Eby, Wayne M.; Singh, Karan P. (2011). "Mathematical modeling of stem cell proliferation". Medical & Biological Engineering & Computing. 49 (3): 253–262. doi:10.1007/s11517-010-0686-y. PMID 20953843.
- Eby, Wayne M.; Tabatabai, Mohammad A. (2014). "Methods in Mathematical Modeling for Stem Cells". Therapeutic Applications in Disease and Injury. Stem Cells and Cancer Stem Cells. 12. pp. 201–217. doi:10.1007/978-94-017-8032-2_18. ISBN 978-94-017-8031-5.
- Wadkin, L. E.; Orozco-Fuentes, S.; Neganova, I.; Lako, M.; Shukurov, A.; Parker, N. G. (2020). "The recent advances in the mathematical modelling of human pluripotent stem cells". SN Applied Sciences. 2 (2). doi:10.1007/s42452-020-2070-3.
- Stem Cells and Cancer Stem Cells, Volume 12. Stem Cells and Cancer Stem Cells. 12. 2014. doi:10.1007/978-94-017-8032-2. ISBN 978-94-017-8031-5.
- Tabatabai, Mohammad A.; Kengwoung-Keumo, Jean-Jacques; Eby, Wayne M.; Bae, Sejong; Guemmegne, Juliette T.; Manne, Upender; Fouad, Mona; Partridge, Edward E.; Singh, Karan P. (2014). "Disparities in Cervical Cancer Mortality Rates as Determined by the Longitudinal Hyperbolastic Mixed-Effects Type II Model". PLOS ONE. 9 (9): e107242. Bibcode:2014PLoSO...9j7242T. doi:10.1371/journal.pone.0107242. PMC 4167327. PMID 25226583.
- Veríssimo, André; Paixão, Laura; Neves, Ana; Vinga, Susana (2013). "BGFit: Management and automated fitting of biological growth curves". BMC Bioinformatics. 14: 283. doi:10.1186/1471-2105-14-283. PMC 3848918. PMID 24067087.
- Tabatabai, M. A.; Eby, W. M.; Bae, S.; Singh, K. P. (2013). "A flexible multivariable model for Phytoplankton growth". Mathematical Biosciences and Engineering. 10 (3): 913–923. doi:10.3934/mbe.2013.10.913. PMID 23906155.
- Eby, Wayne M.; Oyamakin, Samuel O.; Chukwu, Angela U. (2017). "A new nonlinear model applied to the height-DBH relationship in Gmelina arborea". Forest Ecology and Management. 397: 139–149. doi:10.1016/j.foreco.2017.04.015.
- Tabatabai, M. A.; Eby, W. M.; Bae, S.; Singh, K. P. (2013). "A flexible multivariable model for Phytoplankton growth". Mathematical Biosciences and Engineering. 10 (3): 913–923. doi:10.3934/mbe.2013.10.913. PMID 23906155.
- Majid, Shahid; Salih, Mohammad; Wasaya, Rozina; Jafri, Wasim (2008). "Predictors of gastrointestinal lesions on endoscopy in iron deficiency anemia without gastrointestinal symptoms". BMC Gastroenterology. 8: 52. doi:10.1186/1471-230X-8-52. PMC 2613391. PMID 18992171.
- Timmerman, Dirk; Testa, Antonia C.; Bourne, Tom; Ferrazzi, Enrico; Ameye, Lieveke; Konstantinovic, Maja L.; Van Calster, Ben; Collins, William P.; Vergote, Ignace; Van Huffel, Sabine; Valentin, Lil (2005). "Logistic Regression Model to Distinguish Between the Benign and Malignant Adnexal Mass Before Surgery: A Multicenter Study by the International Ovarian Tumor Analysis Group". Journal of Clinical Oncology. 23 (34): 8794–8801. doi:10.1200/JCO.2005.01.7632. PMID 16314639.