Beta bulge loop
Beta bulge loops are commonly occurring motifs in proteins and polypeptides consisting of five to six amino acids.[1][2][3] There are two types: type 1, which is a pentapeptide; and type 2, with six amino acids. They are regarded as a type of beta bulge, and have the alternative name of type G1 beta bulge. Compared to other beta bulges, beta bulge loops give rise to chain reversal such that they often occur at the loop ends of beta hairpins; hairpins of this sort can be described as 3:5 (for a type 1 β bulge loop) or 4:6 (for type 2).[4] Two websites are available for finding and examining β bulge loops in proteins, Motivated Proteins: [5] and PDBeMotif: .[6]
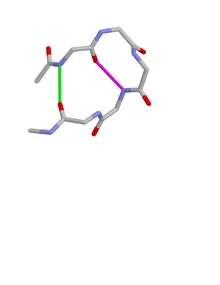
Type I beta bulge loops have two characteristic inter-main-chain hydrogen bonds. One is between the CO of residue i and the NH of residue i+3 (a β-turn); the other is between the CO of residue i+4 and the NH of residue i.
Type 2 beta bulge loops have two characteristic inter-main-chain hydrogen bonds. One is between the CO of residue i and the NH of residue i+4 (an α-turn); the other is between the CO of residue i+5 and the NH of residue i.
Beta bulge loops often have an aspartate, asparagine, serine or threonine at residue i, together with a nest (protein structural motif) at residues i+2 to i+4 (type 1) or residues i+3 to i+5 (type 2), with the side chain oxygen binding to the main chain NH groups of the nest.[7] Site-directed mutagenesis of asx residues within a protein's β bulge loops has been described, showing that the side chain of an asx residue at various alternative positions within a β bulge loop binds to the nest and thereby helps stabilize the loop.[8]
References
- Milner-White, EJ (1987). "Beta bulges within loops as recurring features of protein structure". Biochim Biophys Acta. 911 (2): 261–265. doi:10.1016/0167-4838(87)90017-3. PMID 3801498.
- Chan, EAW; Hutchinson EG (1993). "Identification, classification and analysis of β-bulges in proteins". Protein Science. 2 (10): 1574–1590. doi:10.1002/pro.5560021004. PMC 2142268. PMID 8251933.
- Blandl, T; Cochran AG (2003). "Turn Stability in β-hairpin peptides. Investigation of peptides containing 3:5 type 1 G1 Bulge turns". Protein Science. 12 (2): 237–247. doi:10.1110/ps.0228603. PMC 2312432. PMID 12538887.
- Sibanda, BL; Blundell TL; Thornton JM (1989). "Conformation of β-hairpins in protein structures. A systematic classification with applications to modeling". Journal of Molecular Biology. 206 (4): 759–777. doi:10.1016/0022-2836(89)90583-4. PMID 2500530.
- Leader, DP; Milner-White EJ (2009). "Motivated Proteins: A web application for studying small three-dimensional protein motifs". BMC Bioinformatics. 10 (1): 60. doi:10.1186/1471-2105-10-60. PMC 2651126. PMID 19210785.
- Golovin, A; Henrick K (2008). "MSDmotif: exploring protein sites and motifs". BMC Bioinformatics. 9 (1): 312. doi:10.1186/1471-2105-9-312. PMC 2491636. PMID 18637174.
- Afzal, AM; Al-Shubailly F (2014). "Bridging of anions by hydrogen bonds in nest motifs and its significance for Schellman motifs and other larger motifs within proteins". Proteins. 82 (11): 3023–3031. doi:10.1002/prot.24663. PMID 25132631.
- Lee, J; Dubey VK; Longo LM; Blaber M (2008). "A logical OR redundancy within the Asx-Pro-Asx-Gly β-turn motif". Journal of Molecular Biology. 377 (4): 1251–1264. doi:10.1016/j.jmb.2008.01.055. PMID 18308335.