Retrotransposon
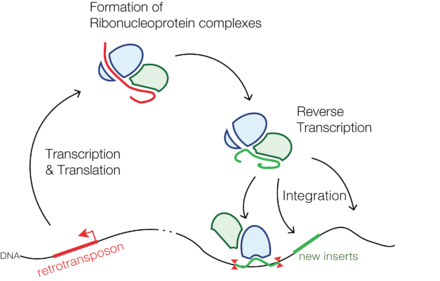
Retrotransposons (also called transposons via RNA intermediates) are genetic elements that can amplify themselves in a genome and are ubiquitous components of the DNA of many eukaryotic organisms. These DNA sequences use a "copy-and-paste" mechanism, whereby they are first transcribed into RNA, then converted back into identical DNA sequences using reverse transcription, and these sequences are then inserted into the genome at target sites.
Retrotransposons form one of the two subclasses of transposons, where the others are DNA transposons, which does not involve an RNA intermediate.
Retrotransposons are particularly abundant in plants, where they are often a principal component of nuclear DNA. In maize, 49–78% of the genome is made up of retrotransposons.[1] In wheat, about 90% of the genome consists of repeated sequences and 68% of transposable elements.[2] In mammals, almost half the genome (45% to 48%) is transposons or remnants of transposons. Around 42% of the human genome is made up of retrotransposons, while DNA transposons account for about 2–3%.[3]
Biological activity
The retrotransposons' replicative mode of transposition by means of an RNA intermediate rapidly increases the copy numbers of elements and thereby can increase genome size. Like DNA transposable elements (class II transposons), retrotransposons can induce mutations by inserting near or within genes. Furthermore, retrotransposon-induced mutations are relatively stable, because the sequence at the insertion site is retained as they transpose via the replication mechanism.
Retrotransposons copy themselves to RNA and then back to DNA that may integrate back to the genome. The second step of forming DNA may be carried out by a reverse transcriptase, which the retrotransposon encodes.[4] Transposition and survival of retrotransposons within the host genome are possibly regulated both by retrotransposon- and host-encoded factors, to avoid deleterious effects on host and retrotransposon as well. The understanding of how retrotransposons and their hosts' genomes have co-evolved mechanisms to regulate transposition, insertion specificities, and mutational outcomes in order to optimize each other's survival is still in its infancy.
Because of accumulated mutations, most retrotransposons are no longer able to retrotranspose.
Types
Retrotransposons, also known as class I transposable elements, consist of two subclasses, the long terminal repeat (LTR-retrotransposons) and the non-LTR retrotransposons. Classification into these subclasses is based on the phylogeny of the reverse transcriptase,[5] which goes in line with structural differences, such as presence/absence of long terminal repeats as well as number and types of open reading frames, encoding domains and target site duplication lengths.
LTR retrotransposons
LTR retrotransposons have direct LTRs that range from ~100 bp to over 5 kb in size. LTR retrotransposons are further sub-classified into the Ty1-copia-like (Pseudoviridae), Ty3-gypsy-like (Metaviridae), and BEL-Pao-like groups based on both their degree of sequence similarity and the order of encoded gene products. Ty1-copia and Ty3-gypsy groups of retrotransposons are commonly found in high copy number (up to a few million copies per haploid nucleus) in animals, fungi, protista, and plants genomes. BEL-Pao like elements have so far only been found in animals.[6][7]
Although retroviruses are often classified separately, they share many features with LTR retrotransposons. A major difference with Ty1-copia and Ty3-gypsy retrotransposons is that retroviruses have an envelope protein (ENV). A retrovirus can be transformed into an LTR retrotransposon through inactivation or deletion of the domains that enable extracellular mobility. If such a retrovirus infects and subsequently inserts itself in the genome in germ line cells, it may become transmitted vertically and become an Endogenous Retrovirus (ERV).[7] Endogenous retroviruses make up about 8% of the human genome and approximately 10% of the mouse genome.[8]
In plant genomes, LTR retrotransposons are the major repetitive sequence class, e.g. able to constitute more than 75% of the maize genome.[9]
Endogenous retroviruses (ERV)
Endogenous retroviruses are an important type of LTR retrotransposon in mammals, including in humans where the Human ERVs make up 8% of the genome.

Non-LTR retrotransposons
Non-LTR retrotransposons consist of two sub-types, long interspersed elements (LINEs) and short interspersed elements (SINEs). They can also be found in high copy numbers, as shown in the plant species.[11] Non-long terminal repeat (LTR) retroposons are widespread in eukaryotic genomes. LINEs possess two ORFs, which encode all the functions needed for retrotransposition. These functions include reverse transcriptase and endonuclease activities, in addition to a nucleic acid-binding property needed to form a ribonucleoprotein particle.[12] SINEs, on the other hand, co-opt the LINE machinery and function as nonautonomous retroelements. While historically viewed as "junk DNA", recent research suggests that, in some rare cases, both LINEs and SINEs were incorporated into novel genes so as to evolve new functionality.[13][14]
LINEs
Long INterspersed Elements[15] (LINE) are a group of genetic elements that are found in large numbers in eukaryotic genomes, comprising 17% of the human genome (99.9% of which is no longer capable of retrotransposition, and therefore considered "dead" or inactive).[16] Among the LINE, there are several subgroups, such as L1, L2 and L3. Human coding L1 begin with an untranslated region (UTR) that includes an RNA polymerase II promoter, two non-overlapping open reading frames (ORF1 and ORF2), and ends with another UTR.[16] Recently, a new open reading frame in the 5' end of the LINE elements has been identified in the reverse strand. It is shown to be transcribed and endogenous proteins are observed. The name ORF0 is coined due to its position with respect to ORF1 and ORF2.[17] ORF1 encodes an RNA binding protein and ORF2 encodes a protein having an endonuclease (e.g. RNase H) as well as a reverse transcriptase. The reverse transcriptase has a higher specificity for the LINE RNA than other RNA, and makes a DNA copy of the RNA that can be integrated into the genome at a new site.[18] The endonuclease encoded by non-LTR retroposons may be AP (Apurinic/Pyrimidinic) type or REL (Restriction Endonuclease Like) type. Elements in the R2 group have REL type endonuclease, which shows site specificity in insertion.[19]
The 5' UTR contains the promoter sequence, while the 3' UTR contains a polyadenylation signal (AATAAA) and a poly-A tail.[20] Because LINEs (and other class I transposons, e.g. LTR retrotransposons and SINEs) move by copying themselves (instead of moving by a cut and paste like mechanism, as class II transposons do), they enlarge the genome. The human genome, for example, contains about 500,000 LINEs, which is roughly 17% of the genome.[21] Of these, approximately 7,000 are full-length, a small subset of which are capable of retrotransposition.[22][23]
Specific LINE-1 retroposons in the human genome are transcribed and the associated LINE-1 RNAs are tightly bound to nucleosomes and essential in the establishment of local chromatin environment.[24]
SINEs
Short interspersed nuclear elements[15] (SINEs) are a group of non-LTR (long terminal repeat) and non-autonomous retrotransposons.
SINEs are the only TEs that are non- autonomous by nature, meaning that they did not evolve from autonomous elements. They are small (80- 500 bases[25])) and rely in trans on functional LINEs for their replication, but their evolutionary origin is very distinct. SINEs can be found in very diverse eukaryotes, but they have only accumulated to impressive amount in mammals, where they represent between 5 and 15% of the genome with millions of copies.
Structure and propagation
SINEs typically possess a “head” with an RNA pol III promoter that enables autonomous transcription, and a body of various composition[26]. SINEs are postulated to originate from the accidental retrotransposition of various RNA pol III transcripts, and have appeared separately numerous times in evolution history. The type of RNA pol III promoter defines the different superfamilies and reveal their origin: tRNA, 5S ribosomal RNA or signal recognition particle 7SL RNA.
SINEs do not encode a functional reverse transcriptase protein and rely on other mobile elements for the transposition, especially LINEs[27]. SINE RNAs form a complex with LINE ORF2 proteins and are inserted into the genome by target primed reverse transcription, creating short TSDs upon insertion. Some SINE families are thought to rely on specific LINEs for their replication, while others seem to be more generalist.
Alu and B1 elements, with their 1.1 million and 650,000 copies in the human and mouse genomes, respectively, harbor a 7SL promoter. The 350,000 copies of B2 SINEs in the mouse are on the other hand tRNA-related.
Types
Alu and B1 elements, with their 1.1 million and 650,000 copies in the human and mouse genomes, respectively, harbor a 7SL promoter.
The 350,000 copies of B2 SINEs in the mouse are on the other hand tRNA- related.
The most common SINE in primates is Alu. Alu elements are approximately 350 base pairs long, do not contain any coding sequences, and can be recognized by the restriction enzyme AluI (hence the name). The distribution of these elements has been implicated in some genetic diseases and cancers.
Hominid genomes contain also original elements termed SVA. They are composite transposons formed by the fusion of a SINE-R and an Alu, separated by a variable number of tandems repeats. Less than 3kb in length and apparently mobilized using LINE1 machinery, they are around 2500-3000 copies in human or gorilla genomes, and less than 1000 in orangutan. SVA are one of the youngest transposable element in great apes genome and among the most active and polymorphic in the human population.
See also
- Transposon
- Endogenous retrovirus
- Paleogenetics
- Paleovirology
- Insertion sequences
- Copy-number variation
- Genomic organization
- Interspersed repeat
- Retrotransposon markers, a powerful method of reconstructing phylogenies.
- RetrOryza
References
- ↑ SanMiguel P, Bennetzen JL (1998). "Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotranposons" (PDF). Annals of Botany. 82 (Suppl A): 37–44. doi:10.1006/anbo.1998.0746.
- ↑ Li W, Zhang P, Fellers JP, Friebe B, Gill BS (November 2004). "Sequence composition, organization, and evolution of the core Triticeae genome". Plant J. 40 (4): 500–11. doi:10.1111/j.1365-313X.2004.02228.x. PMID 15500466.
- ↑ Lander ES, Linton LM, Birren B, et al. (February 2001). "Initial sequencing and analysis of the human genome". Nature. 409 (6822): 860–921. doi:10.1038/35057062. PMID 11237011.
- ↑ Dombroski BA, Feng Q, Mathias SL, et al. (July 1994). "An in vivo assay for the reverse transcriptase of human retrotransposon L1 in Saccharomyces cerevisiae". Mol. Cell. Biol. 14 (7): 4485–92. doi:10.1128/mcb.14.7.4485. PMC 358820. PMID 7516468.
- ↑ Xiong, Y; Eickbush, TH (October 1990). "Origin and evolution of retroelements based upon their reverse transcriptase sequences". The EMBO Journal. 9 (10): 3353–62. PMC 552073. PMID 1698615.
- ↑ Copeland CS, Mann VH, Morales ME, Kalinna BH, Brindley PJ (2005). "The Sinbad retrotransposon from the genome of the human blood fluke, Schistosoma mansoni, and the distribution of related Pao-like elements". BMC Evol. Biol. 5 (1): 20. doi:10.1186/1471-2148-5-20. PMC 554778. PMID 15725362.
- 1 2 Wicker T, Sabot F, Hua-Van A, et al. (December 2007). "A unified classification system for eukaryotic transposable elements". Nat. Rev. Genet. 8 (12): 973–82. doi:10.1038/nrg2165. PMID 17984973.
- ↑ McCarthy EM, McDonald JF (2004). "Long terminal repeat retrotransposons of Mus musculus". Genome Biol. 5 (3): R14. doi:10.1186/gb-2004-5-3-r14. PMC 395764. PMID 15003117.
- ↑ Baucom, RS; Estill, JC; Chaparro, C; Upshaw, N; Jogi, A; Deragon, JM; Westerman, RP; Sanmiguel, PJ; Bennetzen, JL (November 2009). "Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome". PLoS Genetics. 5 (11): e1000732. doi:10.1371/journal.pgen.1000732. PMC 2774510. PMID 19936065.
- ↑ "Transposon regulation upon dynamic loss of DNA methylation (PDF Download Available)". ResearchGate. doi:10.13140/rg.2.2.18747.21286.
- ↑ Schmidt, Thomas (1999-08-01). "LINEs, SINEs and repetitive DNA: non-LTR retrotransposons in plant genomes". Plant Molecular Biology. 40 (6): 903–910. doi:10.1023/A:1006212929794. ISSN 0167-4412.
- ↑ Yadav, VP; Mandal, PK; Rao, DN; Bhattacharya, S (December 2009). "Characterization of the restriction enzyme-like endonuclease encoded by the Entamoeba histolytica non-long terminal repeat retroposon EhLINE1". The FEBS Journal. 276 (23): 7070–82. doi:10.1111/j.1742-4658.2009.07419.x. PMID 19878305.
- ↑ Santangelo AM, de Souza FS, Franchini LF, Bumaschny VF, Low MJ, Rubinstein M (October 2007). "Ancient Exaptation of a CORE-SINE Retroposon into a Highly Conserved Mammalian Neuronal Enhancer of the Proopiomelanocortin Gene". PLoS Genetics. Public Library of Science. 3 (10): 1813–26. doi:10.1371/journal.pgen.0030166. PMC 2000970. PMID 17922573. Retrieved 2007-12-31.
- ↑ Liang, Kung-Hao; Yeh, Chau-Ting (2013). "A gene expression restriction network mediated by sense and antisense Alu sequences located on protein-coding messenger RNAs". BMC Genomics. 14: 325. doi:10.1186/1471-2164-14-325. PMC 3655826. PMID 23663499. Retrieved 2013-05-11.
- 1 2 Singer MF (March 1982). "SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes". Cell. 28 (3): 433–4. doi:10.1016/0092-8674(82)90194-5. PMID 6280868.
- 1 2 Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, Athanikar JN, Hasnaoui M, Bucheton A, Moran JV, Gilbert N (October 7, 2010). "Characterization of LINE-1 ribonucleoprotein particles". PLOS Genetics. 6 (10): e1001150. doi:10.1371/journal.pgen.1001150. PMC 2951350. PMID 20949108.
- ↑ Denli, AM; Narvaiza, I; Kerman, BE; Pena, M; Benner, C; Marchetto, MC; Diedrich, JK; Aslanian, A; Ma, J; Moresco, JJ; Moore, L; Hunter, T; Saghatelian, A; Gage, FH (22 October 2015). "Primate-Specific ORF0 Contributes to Retrotransposon-Mediated Diversity". Cell. 163 (3): 583–93. doi:10.1016/j.cell.2015.09.025. PMID 26496605.
- ↑ Ohshima K, Okada N (2005). "SINEs and LINEs: symbionts of eukaryotic genomes with a common tail". Cytogenet. Genome Res. 110 (1–4): 475–90. doi:10.1159/000084981. PMID 16093701.
- ↑ Yadav, VP; Mandal, PK; Rao, DN; Bhattacharya, S (December 2009). "Characterization of the restriction enzyme-like endonuclease encoded by the Entamoeba histolytica non-long terminal repeat retrotransposon EhLINE1". The FEBS Journal. 276 (23): 7070–82. doi:10.1111/j.1742-4658.2009.07419.x. PMID 19878305.
- ↑ Deininger PL, Batzer MA (October 2002). "Mammalian retroelements". Genome Res. 12 (10): 1455–65. doi:10.1101/gr.282402. PMID 12368238.
- ↑ Richard Cordaux; Mark Batzer (October 2009). "The impact of retrotransposons on human genome evolution". Nature Reviews Genetics. 10 (10): 691–703. doi:10.1038/nrg2640. PMC 2884099. PMID 19763152.
- ↑ Griffiths, Anthony J. (2008). Introduction to genetic analysis (9th ed.). New York: W.H. Freeman. p. 505. ISBN 0-7167-6887-9.
- ↑ Rangwala S, Kazazian HH (2009). "Many LINE1 elements contribute to the transcriptome of human somatic cells". Genome Biology. 10 (9): R100. doi:10.1186/gb-2009-10-9-r100. PMC 2768975. PMID 19772661.
- ↑ Chueh, A.C.; Northrop, Emma L.; Brettingham-Moore, Kate H.; Choo, K. H. Andy; Wong, Lee H. (Jan 2009). Bickmore, Wendy A., ed. "LINE Retrotransposon RNA Is an Essential Structural and Functional Epigenetic Component of a Core Neocentromeric Chromatin". PLoS Genetics. 5 (1): e1000354. doi:10.1371/journal.pgen.1000354. PMC 2625447. PMID 19180186.
- ↑ Stansfield, William D.; King, Robert C. (1997). A dictionary of genetics (5th ed.). Oxford [Oxfordshire]: Oxford University Press. ISBN 0-19-509441-7.
- ↑ KRAMEROV, D; VASSETZKY, N. "Short Retroposons in Eukaryotic Genomes". International Review of Cytology. 247: 165–221. doi:10.1016/s0074-7696(05)47004-7.
- ↑ Dewannieux, Marie; Esnault, Cécile; Heidmann, Thierry. "LINE-mediated retrotransposition of marked Alu sequences". Nature Genetics. 35 (1): 41–48. doi:10.1038/ng1223.