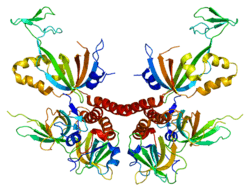Replication protein A3
Replication protein A 14 kDa subunit is a protein that in humans is encoded by the RPA3 gene.[5][6]
Interactions
RPA3 has been shown to interact with replication protein A1[7][8] and replication protein A2.[7][8]
See also
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000106399 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000012483 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Umbricht CB, Erdile LF, Jabs EW, Kelly TJ (Mar 1993). "Cloning, overexpression, and genomic mapping of the 14-kDa subunit of human replication protein A". The Journal of Biological Chemistry. 268 (9): 6131–8. PMID 8454588.
- ↑ "Entrez Gene: RPA3 replication protein A3, 14kDa".
- 1 2 Bochkareva E, Frappier L, Edwards AM, Bochkarev A (Feb 1998). "The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain". The Journal of Biological Chemistry. 273 (7): 3932–6. doi:10.1074/jbc.273.7.3932. PMID 9461578.
- 1 2 Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A (Apr 2002). "Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA". The EMBO Journal. 21 (7): 1855–63. doi:10.1093/emboj/21.7.1855. PMC 125950. PMID 11927569.
Further reading
- Iftode C, Daniely Y, Borowiec JA (1999). "Replication protein A (RPA): the eukaryotic SSB". Critical Reviews in Biochemistry and Molecular Biology. 34 (3): 141–80. doi:10.1080/10409239991209255. PMID 10473346.
- Keshav KF, Chen C, Dutta A (Jun 1995). "Rpa4, a homolog of the 34-kilodalton subunit of the replication protein A complex". Molecular and Cellular Biology. 15 (6): 3119–28. doi:10.1128/mcb.15.6.3119. PMC 230543. PMID 7760808.
- Umbricht CB, Griffin CA, Hawkins AL, Grzeschik KH, O'Connell P, Leach R, Green ED, Kelly TJ (Mar 1994). "High-resolution genomic mapping of the three human replication protein A genes (RPA1, RPA2, and RPA3)". Genomics. 20 (2): 249–57. doi:10.1006/geno.1994.1161. PMID 8020972.
- Amacker M, Hottiger M, Mossi R, Hübscher U (Mar 1997). "HIV-1 nucleocapsid protein and replication protein A influence the strand displacement DNA synthesis of lentiviral reverse transcriptase". AIDS. 11 (4): 534–6. PMID 9084803.
- Bochkareva E, Frappier L, Edwards AM, Bochkarev A (Feb 1998). "The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain". The Journal of Biological Chemistry. 273 (7): 3932–6. doi:10.1074/jbc.273.7.3932. PMID 9461578.
- "Toward a complete human genome sequence". Genome Research. 8 (11): 1097–108. Nov 1998. doi:10.1101/gr.8.11.1097. PMID 9847074.
- Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ (Oct 2000). "Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA". Cell. 103 (3): 449–56. doi:10.1016/S0092-8674(00)00136-7. PMID 11081631.
- Habel JE, Ohren JF, Borgstahl GE (Feb 2001). "Dynamic light-scattering analysis of full-length human RPA14/32 dimer: purification, crystallization and self-association". Acta Crystallographica Section D. 57 (Pt 2): 254–9. doi:10.1107/S0907444900015225. PMID 11173472.
- Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A (Apr 2002). "Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA". The EMBO Journal. 21 (7): 1855–63. doi:10.1093/emboj/21.7.1855. PMC 125950. PMID 11927569.
- Kneissl M, Pütter V, Szalay AA, Grummt F (Mar 2003). "Interaction and assembly of murine pre-replicative complex proteins in yeast and mouse cells". Journal of Molecular Biology. 327 (1): 111–28. doi:10.1016/S0022-2836(03)00079-2. PMID 12614612.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (Oct 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. doi:10.1038/nature04209. PMID 16189514.
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, Vasilescu J, Abu-Farha M, Lambert JP, Duewel HS, Stewart II, Kuehl B, Hogue K, Colwill K, Gladwish K, Muskat B, Kinach R, Adams SL, Moran MF, Morin GB, Topaloglou T, Figeys D (2007). "Large-scale mapping of human protein-protein interactions by mass spectrometry". Molecular Systems Biology. 3 (1): 89. doi:10.1038/msb4100134. PMC 1847948. PMID 17353931.
- Sankaranarayanan P, Schomay TE, Aiello KA, Alter O (April 2015). "Tensor GSVD of patient- and platform-matched tumor and normal DNA copy-number profiles uncovers chromosome arm-wide patterns of tumor-exclusive platform-consistent alterations encoding for cell transformation and predicting ovarian cancer survival". PLOS ONE. 10 (4): e0121396. doi:10.1371/journal.pone.0121396. PMC 4398562. PMID 25875127. AAAS EurekAlert! Press Release and NAE Podcast Feature.
This article is issued from
Wikipedia.
The text is licensed under Creative Commons - Attribution - Sharealike.
Additional terms may apply for the media files.