Molecular machine
| Part of a series of articles on |
| Molecular nanotechnology |
|---|
|
A molecular machine, nanite, or nanomachine,[1] refers to any discrete number of molecular components that produce quasi-mechanical movements (output) in response to specific stimuli (input).[2] In biology, macromolecular machines frequently perform tasks essential for life such as DNA replication and ATP synthesis. The expression is often more generally applied to molecules that simply mimic functions that occur at the macroscopic level. The term is also common in nanotechnology where a number of highly complex molecular machines have been proposed that are aimed at the goal of constructing a molecular assembler.
For the last several decades, chemists and physicists alike have attempted, with varying degrees of success, to miniaturize machines found in the macroscopic world. Molecular machines research is currently at the forefront with the 2016 Nobel Prize in Chemistry being awarded to Jean-Pierre Sauvage, Sir J. Fraser Stoddart and Bernard L. Feringa for the design and synthesis of molecular machines.[3][4]
Types
Molecular machines can be divided into two broad categories; synthetic and biological. In general, synthetic molecular machines refer to molecules that are artificially designed and synthesized whereas biological molecular machines can commonly be found in nature.[5]
Synthetic
A wide variety of rather simple molecular machines have been synthesized by chemists. They can consist of a single molecule; however, they are often constructed for mechanically-interlocked molecular architectures, such as rotaxanes and catenanes. Carbon nanotube nanomotors have also been produced.[6]
- Molecular motors are molecules that are capable of unidirectional rotation motion powered by external energy input. A number of molecular machines have been synthesized powered by light or reaction with other molecules.[7][8][9][10]
- A molecular propeller is a molecule that can propel fluids when rotated, due to its special shape that is designed in analogy to macroscopic propellers. It has several molecular-scale blades attached at a certain pitch angle around the circumference of a nanoscale shaft. Also see molecular gyroscope.
- A molecular switch is a molecule that can be reversibly shifted between two or more stable states.[11] The molecules may be shifted between the states in response to changes in pH, light, temperature, an electric current, microenvironment, or the presence of a ligand.
- A molecular shuttle is a molecule capable of shuttling molecules or ions from one location to another. A common molecular shuttle consists of a rotaxane where the macrocycle can move between two sites or stations along the dumbbell backbone.[12]
- A molecular balance[13][14] is a molecule that can interconvert between two and more conformational or configurational states in response to the dynamic of multiple intra- and intermolecular driving forces, such as hydrogen bonding, solvophobic/hydrophobic effects,[15] π interactions,[16] and steric and dispersion interactions.[17]
- Molecular tweezers are host molecules capable of holding items between their two arms. The open cavity of the molecular tweezers binds items using non-covalent bonding including hydrogen bonding, metal coordination, hydrophobic forces, van der Waals forces, π interactions, or electrostatic effects. Examples of molecular tweezers have been reported that are constructed from DNA and are considered DNA machines.
- A molecular sensor is a molecule that interacts with an analyte to produce a detectable change.[18] Molecular sensors combine molecular recognition with some form of reporter, so the presence of the item can be observed.
- A molecular logic gate is a molecule that performs a logical operation on one or more logic inputs and produces a single logic output. Unlike a molecular sensor, the molecular logic gate will only output when a particular combination of inputs are present.
- A molecular assembler is a molecular machine able to guide chemical reactions by positioning reactive molecules with precision.[20][21][22][23][24]
- A molecular hinge is a molecule that can be selectively switched from one configuration to another in a reversible fashion.[11] Such configurations must have distinguishable geometries, for instance, Cis or Trans isomers[25] of a V-shape[26] molecule. Azo compounds perform Cis–trans isomerism upon receiving UV-Vis light.[11]
Biological
The most complex macromolecular machines are found within cells, often in the form of multi-protein complexes.[27] Some biological machines are motor proteins, such as myosin, which is responsible for muscle contraction, kinesin, which moves cargo inside cells away from the nucleus along microtubules, and dynein, which moves cargo inside cells towards the nucleus and produces the axonemal beating of motile cilia and flagella. "[I]n effect, the [motile cilium] is a nanomachine composed of perhaps over 600 proteins in molecular complexes, many of which also function independently as nanomachines...Flexible linkers allow the mobile protein domains connected by them to recruit their binding partners and induce long-range allostery via protein domain dynamics. "[1] Other biological machines are responsible for energy production, for example ATP synthase which harnesses energy from proton gradients across membranes to drive a turbine-like motion used to synthesise ATP, the energy currency of a cell.[28] Still other machines are responsible for gene expression, including DNA polymerases for replicating DNA, RNA polymerases for producing mRNA, the spliceosome for removing introns, and the ribosome for synthesising proteins. These machines and their nanoscale dynamics are far more complex than any molecular machines that have yet been artificially constructed.[29]
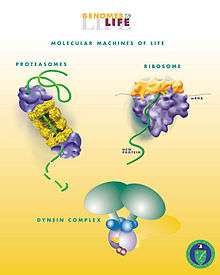
These biological machines might have applications in nanomedicine. For example,[30] they could be used to identify and destroy cancer cells.[31][32] Molecular nanotechnology is a speculative subfield of nanotechnology regarding the possibility of engineering molecular assemblers, biological machines which could re-order matter at a molecular or atomic scale. Nanomedicine would make use of these nanorobots, introduced into the body, to repair or detect damages and infections. Molecular nanotechnology is highly theoretical, seeking to anticipate what inventions nanotechnology might yield and to propose an agenda for future inquiry. The proposed elements of molecular nanotechnology, such as molecular assemblers and nanorobots are far beyond current capabilities.[33][34]
Research
The construction of more complex molecular machines is an active area of theoretical and experimental research. A number of molecules, such as molecular propellers, have been designed, although experimental studies of these molecules are inhibited by the lack of methods to construct these molecules. In this context, theoretical modeling can be extremely useful to understand the self-assembly/disassembly processes of rotaxanes, important for the construction of light-powered molecular machines.[35] This molecular-level knowledge may foster the realization of ever more complex, versatile, and effective molecular machines for the areas of nanotechnology, including molecular assemblers.
Although currently not feasible, some potential applications of molecular machines are transport at the molecular level, manipulation of nanostructures and chemical systems, high density solid-state informational processing and molecular prosthetics.[36] Many fundamental challenges need to be overcome before molecular machines can be used practically such as autonomous operation, complexity of machines, stability in the synthesis of the machines and the working conditions.[5]
In 2018, an international team of researchers, led by researchers from the University of Osaka, Japan, created a molecular machine in which elements of a mechanical ratchet were used. The main advantage of this machine is that it provides movement in only one direction. In addition, some features of the structure of the molecular machine provide the best balance between the produced motion and chemical reactivity of the molecules that make up it, that is a problem in itself.[37]
References
- 1 2 Satir, Peter; Søren T. Christensen (2008-03-26). "Structure and function of mammalian cilia". Histochemistry and Cell Biology. Springer Berlin / Heidelberg. 129 (6): 688. doi:10.1007/s00418-008-0416-9. PMC 2386530. PMID 18365235. 1432-119X. Retrieved 2009-09-11.
- ↑ Ballardini R, Balzani V, Credi A, Gandolfi MT, Venturi M (2001). "Artificial Molecular-Level Machines: Which Energy To Make Them Work?". Acc. Chem. Res. 34 (6): 445–455. doi:10.1021/ar000170g.
- ↑ Staff (5 October 2016). "The Nobel Prize in Chemistry 2016". Nobel Foundation. Retrieved 5 October 2016.
- ↑ Chang, Kenneth; Chan, Sewell (5 October 2016). "3 Makers of 'World's Smallest Machines' Awarded Nobel Prize in Chemistry". New York Times. Retrieved 5 October 2016.
- 1 2 Erbas-Cakmak, Sundus; Leigh, David A.; McTernan, Charlie T.; Nussbaumer, Alina L. (2015). "Artificial Molecular Machines". Chemical Reviews. 115 (18): 10081–10206. doi:10.1021/acs.chemrev.5b00146.
- ↑ Fennimore, A. M.; T.D. Yuzvinsky; Wei-Qiang Han; M. S. Fuhrer; J. Cumings & A. Zettl (2003). "Rotational actuators based on carbon nanotubes". Nature. 424 (6947): 408–410. Bibcode:2003Natur.424..408F. doi:10.1038/nature01823. PMID 12879064.
- ↑ Fletcher, Stephen P.; Dumur, Frédéric; Pollard, Michael M.; Feringa, Ben L. (2005-10-07). "A Reversible, Unidirectional Molecular Rotary Motor Driven by Chemical Energy". Science. 310 (5745): 80–82. Bibcode:2005Sci...310...80F. doi:10.1126/science.1117090. ISSN 0036-8075. PMID 16210531.
- ↑ Perera, U. G. E.; Ample, F.; Kersell, H.; Zhang, Y.; Vives, G.; Echeverria, J.; Grisolia, M.; Rapenne, G.; Joachim, C. (January 2013). "Controlled clockwise and anticlockwise rotational switching of a molecular motor". Nature Nanotechnology. 8 (1): 46–51. Bibcode:2013NatNa...8...46P. doi:10.1038/nnano.2012.218. ISSN 1748-3395.
- ↑ Schliwa, Manfred; Woehlke, Günther (2003-04-17). "Molecular motors". Nature. 422 (6933): 759–765. Bibcode:2003Natur.422..759S. doi:10.1038/nature01601.
- ↑ van Delden, Richard A.; Wiel, Matthijs K. J. ter; Pollard, Michael M.; Vicario, Javier; Koumura, Nagatoshi; Feringa, Ben L. (October 2005). "Unidirectional molecular motor on a gold surface". Nature. 437 (7063): 1337–1340. Bibcode:2005Natur.437.1337V. doi:10.1038/nature04127. ISSN 1476-4687.
- 1 2 3 Kazem-Rostami, Masoud; Moghanian, Amirhossein (2017). "Hünlich base derivatives as photo-responsive Λ-shaped hinges". Organic Chemistry Frontiers. 4: 224–228. doi:10.1039/C6QO00653A.
- ↑ Chatterjee, Manashi N.; Kay, Euan R.; Leigh, David A. (2006-03-01). "Beyond Switches: Ratcheting a Particle Energetically Uphill with a Compartmentalized Molecular Machine". Journal of the American Chemical Society. 128 (12): 4058–4073. doi:10.1021/ja057664z. ISSN 0002-7863.
- ↑ Paliwal, S.; Geib, S.; Wilcox, C. S. (1994-05-01). "Molecular Torsion Balance for Weak Molecular Recognition Forces. Effects of "Tilted-T" Edge-to-Face Aromatic Interactions on Conformational Selection and Solid-State Structure". Journal of the American Chemical Society. 116 (10): 4497–4498. doi:10.1021/ja00089a057. ISSN 0002-7863.
- ↑ Mati, Ioulia K.; Cockroft, Scott L. (2010-10-19). "Molecular balances for quantifying non-covalent interactions". Chemical Society Reviews. 39 (11): 4195. doi:10.1039/B822665M. ISSN 1460-4744.
- ↑ Yang, Lixu; Adam, Catherine; Cockroft, Scott L. (2015-08-19). "Quantifying Solvophobic Effects in Nonpolar Cohesive Interactions". Journal of the American Chemical Society. 137 (32): 10084–10087. doi:10.1021/jacs.5b05736. ISSN 0002-7863.
- ↑ Li, Ping; Zhao, Chen; Smith, Mark D.; Shimizu, Ken D. (2013-06-07). "Comprehensive Experimental Study of N-Heterocyclic π-Stacking Interactions of Neutral and Cationic Pyridines". The Journal of Organic Chemistry. 78 (11): 5303–5313. doi:10.1021/jo400370e. ISSN 0022-3263.
- ↑ Hwang, Jungwun; Li, Ping; Smith, Mark D.; Shimizu, Ken D. (2016-07-04). "Distance-Dependent Attractive and Repulsive Interactions of Bulky Alkyl Groups". Angewandte Chemie International Edition. 55 (28): 8086–8089. doi:10.1002/anie.201602752. ISSN 1521-3773.
- ↑ Cavalcanti A, Shirinzadeh B, Freitas Jr RA, Hogg T (2008). "Nanorobot architecture for medical target identification". Nanotechnology. 19 (1): 015103(15pp). Bibcode:2008Nanot..19a5103C. doi:10.1088/0957-4484/19/01/015103.
- ↑ Org. Chem. Front. 2017, 4 (2), 224-228 https://doi.org/10.1039/c6qo00653a
- ↑ Lewandowski, Bartosz; De Bo, Guillaume; Ward, John W.; Papmeyer, Marcus; Kuschel, Sonja; Aldegunde, María J.; Gramlich, Philipp M. E.; Heckmann, Dominik; Goldup, Stephen M. (2013-01-11). "Sequence-Specific Peptide Synthesis by an Artificial Small-Molecule Machine". Science. 339 (6116): 189–193. Bibcode:2013Sci...339..189L. doi:10.1126/science.1229753. ISSN 0036-8075. PMID 23307739.
- ↑ De Bo, Guillaume; Kuschel, Sonja; Leigh, David A.; Lewandowski, Bartosz; Papmeyer, Marcus; Ward, John W. (2014-04-16). "Efficient Assembly of Threaded Molecular Machines for Sequence-Specific Synthesis". Journal of the American Chemical Society. 136 (15): 5811–5814. doi:10.1021/ja5022415. ISSN 0002-7863.
- ↑ De Bo, Guillaume; Gall, Malcolm A. Y.; Kitching, Matthew O.; Kuschel, Sonja; Leigh, David A.; Tetlow, Daniel J.; Ward, John W. (2017-08-09). "Sequence-Specific β-Peptide Synthesis by a Rotaxane-Based Molecular Machine". Journal of the American Chemical Society. 139 (31): 10875–10879. doi:10.1021/jacs.7b05850. ISSN 0002-7863.
- ↑ Kassem, Salma; Lee, Alan T. L.; Leigh, David A.; Marcos, Vanesa; Palmer, Leoni I.; Pisano, Simone (September 2017). "Stereodivergent synthesis with a programmable molecular machine". Nature. 549 (7672): 374–378. Bibcode:2017Natur.549..374K. doi:10.1038/nature23677. ISSN 1476-4687.
- ↑ De Bo, Guillaume; Gall, Malcolm A. Y.; Kuschel, Sonja; Winter, Julien De; Gerbaux, Pascal; Leigh, David A. (2018-04-02). "An artificial molecular machine that builds an asymmetric catalyst". Nature Nanotechnology. Bibcode:2018NatNa..13..381D. doi:10.1038/s41565-018-0105-3. ISSN 1748-3395.
- ↑ Uznanski, P.; Kryszewski, M.; Thulstrup, E.W. (1991). "Linear dichroism and trans → cis photo-isomerization studies of azobenzene molecules in oriented polyethylene matrix". Eur. Polym. J. 27: 41–43. doi:10.1016/0014-3057(91)90123-6.
- ↑ "Design and synthesis of Ʌ-shaped photoswitchable compounds employing Tröger's base scaffold". Synthesis. 49 (6): 1214–1222. 2017. doi:10.1055/s-0036-1588913.
- ↑ Donald,, Voet, (2011). Biochemistry. Voet, Judith G., (4th ed.). Hoboken, NJ: John Wiley & Sons. ISBN 9780470570951. OCLC 690489261.
- ↑ Kinbara, Kazushi; Aida, Takuzo (2005-04-01). "Toward Intelligent Molecular Machines: Directed Motions of Biological and Artificial Molecules and Assemblies". Chemical Reviews. 105 (4): 1377–1400. doi:10.1021/cr030071r. ISSN 0009-2665.
- ↑ Bu Z, Callaway DJ (2011). "Proteins MOVE! Protein dynamics and long-range allostery in cell signaling". Advances in Protein Chemistry and Structural Biology. Advances in Protein Chemistry and Structural Biology. 83: 163–221. doi:10.1016/B978-0-12-381262-9.00005-7. ISBN 9780123812629. PMID 21570668.
- ↑ Amrute-Nayak, M.; Diensthuber, R. P.; Steffen, W.; Kathmann, D.; Hartmann, F. K.; Fedorov, R.; Urbanke, C.; Manstein, D. J.; Brenner, B.; Tsiavaliaris, G. (2010). "Targeted Optimization of a Protein Nanomachine for Operation in Biohybrid Devices". Angewandte Chemie. 122 (2): 322–326. doi:10.1002/ange.200905200.
- ↑ Patel, G. M.; Patel, G. C.; Patel, R. B.; Patel, J. K.; Patel, M. (2006). "Nanorobot: A versatile tool in nanomedicine". Journal of Drug Targeting. 14 (2): 63–7. doi:10.1080/10611860600612862. PMID 16608733.
- ↑ Balasubramanian, S.; Kagan, D.; Jack Hu, C. M.; Campuzano, S.; Lobo-Castañon, M. J.; Lim, N.; Kang, D. Y.; Zimmerman, M.; Zhang, L.; Wang, J. (2011). "Micromachine-Enabled Capture and Isolation of Cancer Cells in Complex Media". Angewandte Chemie International Edition. 50 (18): 4161–4164. doi:10.1002/anie.201100115. PMC 3119711. PMID 21472835.
- ↑ Freitas, Robert A., Jr.; Havukkala, Ilkka (2005). "Current Status of Nanomedicine and Medical Nanorobotics" (PDF). Journal of Computational and Theoretical Nanoscience. 2 (4): 1–25. Bibcode:2005JCTN....2..471K. doi:10.1166/jctn.2005.001.
- ↑ Nanofactory Collaboration
- ↑ Tabacchi, G.; Silvi, S.; Venturi, M.; Credi, A.; Fois, E. (2016). "Dethreading of a Photoactive Azobenzene-Containing Molecular Axle from a Crown Ether Ring: A Computational Investigation". ChemPhysChem. 17: 1913–1919. doi:10.1002/cphc.201501160.
- ↑ Coskun, Ali; Banaszak, Michal; Astumian, R. Dean; Stoddart, J. Fraser; Grzybowski, Bartosz A. (2011-12-05). "Great expectations: can artificial molecular machines deliver on their promise?". Chem. Soc. Rev. 41 (1): 19–30. doi:10.1039/c1cs15262a. ISSN 1460-4744.
- ↑ "Ratchet Up The Pressure: Molecular Machine Exploits Motion In A Single Direction" ECN Magazine, June 22, 2018