Ancestry-informative marker
In population genetics, an ancestry-informative marker (AIM) is a single-nucleotide polymorphism that exhibits substantially different frequencies between different populations. A set of many AIMs can be used to estimate the proportion of ancestry of an individual derived from each population.
As one example, the Duffy Null allele (FY*0) has a frequency of almost 100% of Sub-Saharan Africans, but occurs very infrequently in populations outside of this region. A person having this allele is thus more likely to have Sub-Saharan African ancestors.
Examining a suite of these markers more or less evenly spaced across the genome is also a cost-effective way to discover novel genes underlying complex diseases in a technique called admixture mapping or mapping by admixture linkage disequilibrium.
There are an estimated 15 million SNP (Single-nucleotide polymorphism) sites (out of roughly 3 billion base pairs, or about 0.4%) from among which AIMs may potentially be selected.[1]
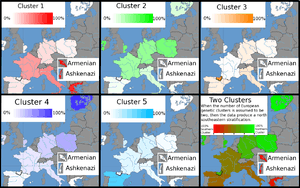
Collections of AIMs have been developed that can estimate the geographical origins of ancestors from within Europe.[2]
North and South Han Chinese ancestry can be distinguished unambiguously using a set of 140 AIMS.[3]
See also
References
- ↑ Elizabeth Pennisi, Human Genetic Variation, Science 21 December 2007: Vol. 318. no. 5858, pp. 1842 - 1843 doi:10.1126/science.318.5858.1842
- ↑ Bauchet M, McEvoy B, Pearson L, Quillen E, Sarkisian T, Hovhannesyan K, Deka R, Bradley D, Shriver M. 2007. Measuring European population stratification with microarray genotype data. American Journal of Human Genetics 80(5): 948-956. doi:10.1086/513477
- ↑ http://www.g3journal.org/content/2/3/339
- General
- Shriver, Mark D. et al., "Skin pigmentation, biogeographical ancestry and admixture mapping," Hum. Genet. 112, 387-399 (2003)
- SNP Science Primer
- dbSNP Summary
- Explanation from DNAPrint Genomics